ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Review Article - Biomedical Research (2018) Volume 29, Issue 17
A review on various attempts for in vitro propagation of human Norovirus
1Institute of Medical and Molecular Biotechnology, Universiti Teknologi MARA, Sungai Buloh, Selangor, Malaysia
2Department of Biochemistry and Molecular Medicine, Universiti Teknologi MARA, Sungai Buloh, Selangor, Malaysia
- *Corresponding Author:
- Mudiana Muhamad
Department of Biochemistry and Molecular Medicine
Universiti Teknologi MARA, Malaysia
Accepted date: August 29, 2018
DOI: 10.4066/biomedicalresearch.29-18-928
Visit for more related articles at Biomedical ResearchHuman noroviruses (HuNoV) are recognized as the primary cause for non-bacterial acute gastroenteritis. The HuNoV outbreak was firstly identified at Norwalk, Ohio and formerly known as ‘winter vomiting disease’ due to the illness frequently strikes during winter season. Infection of HuNoV is estimated to affects all age groups in both developed and developing country, by presenting symptoms such as nausea, vomiting, diarrhea and low-grade fever. There are approximately seven genogroups of noroviruses have been identified in which genogroup II.4 is the dominant HuNoV causing disease to human. Despite their significant impact on the economic and health burden, noroviruses remain to be poorly characterized RNA virus due to the lack of reproducible in vitro culture system for the HuNoV propagation, thus hampering understanding of its pathogenesis. Several approaches including the cultivation trials and the discovery of murine Norovirus (MuNoV) as the model system have expanded our knowledge in Norovirus biology and life cycle. However, to date there are no reliable culture system for HuNoV has been reported. The development of 3D co-culture system has shown a promising future as the improvised system that can be utilized not only for HuNoV cultivation, but also to other viruses. In this review, various attempts to cultivate the HuNoV are discussed to emphasise the future strategy in obtaining reproducible culture system. This review aims to find the gaps in the search of a reliable and reproducible propagation system thus enhancing the understanding on HuNoV biology and pathogenesis for future development of accurate detection method and vaccine production.
Keywords
Noroviruses, Human Norovirus, Norovirus replication, In vitro propagation.
Introduction
Norovirus, a positive strand RNA genome virus belonging to Caliciviridae family has been recognized as the main causative nonbacterial pathogen for acute gastroenteritis. The outbreak of Norovirus was reported to be responsible for over 685 million cases worldwide with an estimation of 21 million cases each year in the US, as recorded by the US Centers for Disease Control and Prevention (CDC) [1,2]. The morbidity symptoms of gastroenteritis caused by human Norovirus (HuNoV) among pediatrics and adult patients include vomiting, watery diarrhea, nausea and mild fever [3]. The virus was initially reported by Zahorsky as ‘winter vomiting disease’ where the prevalence of disease was found to increase during the winter seasons [4,5]. The Norovirus was previously known as “Norwalk virus” signifying the earliest outbreak that led to the Norovirus identification in Norwalk, Ohio, USA. This round structured virus possesses genome which is approximately 7.6 kb in length and classified into several genogroups [6,7]. To date, seven genogroups with over 30 strains have been identified whereby genogroups I, II and IV (GI, GII and GIV) are the major human-related infectious groups while GIII and GV consists of bovine and murine Norovirus, respectively [8,9].
The HuNoV are mainly transmitted via oral route through contaminated food, water or environment targeting human’s gastrointestinal tract. It has been postulated that upon entering the host cells, the noroviruses bind to specific receptors being expressed on the surface of human gastrointestinal epithelium, most likely the M cells. These are specialized epithelial cells comprising of approximately 10% of the human gastrointestinal tract.
The clinical importance of noroviruses attracted global researchers to enhance their knowledge on the virus characteristic as well as the pathogenicity and host immune activity prior to viral entry. However, challenges in cultivation of the HuNoV in vitro limits further understanding of the virus pathogenesis. Furthermore, lack of HuNoV fresh isolates hampers multi-trial viral RNA replication study. This review will cover several fundamental aspects need to be considered for the successful and reproducible in vitro cultivation of HuNoV, thus, filling in the fundamental gaps in HuNoV life cycle. In addition, the succession of HuNoV cultivation will provide the conclusive knowledge on HuNoV molecular biology thus will be beneficial for the manipulation of its cellular mechanism during infection. This will lead to the development of a detection system specific for the viral proteins essential for HuNoV replication in the host cells. More importantly, a successful culture of the HuNoV will be a crucial system to control the virus growth thus providing opportunities for therapeutics and also vaccine development.
Norovirus Replication
Norovirus genome structure
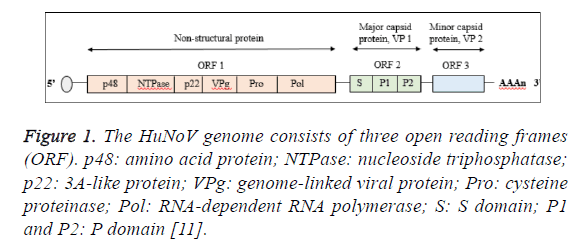
The virus genome consists of three open reading frames (ORF) encoded for eight different viral proteins designated as nonstructural protein (ORF1), major capsid protein (VP1; ORF2) and minor capsid protein (VP2; ORF3) [10]. The ORF1 contains six non-structural proteins which are amino terminal protein (p48), nucleoside triphosphatase (NTPase), 3A-like protein (p22), genome-linked viral protein (VPg), cycteine proteinase (Pro) and RNA-dependent RNA polymerase (Pol) where by two structural proteins are encoded by ORF2 and ORF3 (Figure 1) [11]. The VP1 protein forms the viral capsid via the self -assembly of 90 dimers consisted of N-terminal shell (S) domain accounted for maintaining the icosahedral scaffold prior to extension by protruding (P) domain [12]. The P domain comprise of P1 subdomain which is closely located to the S domain and P2 subdomain. However, the P2 subdomain varied between strains, making the P domain as the vital contributor in the genetic diversity of Norovirus [13]. Detail genotypic and nomenclature analysis of Norovirus revealed more complex classification of their genotypes [14].
Figure 1: The HuNoV genome consists of three open reading frames (ORF). p48: amino acid protein; NTPase: nucleoside triphosphatase; p22: 3A-like protein; VPg: genome-linked viral protein; Pro: cysteine proteinase; Pol: RNA-dependent RNA polymerase; S: S domain; P1 and P2: P domain [11].
The HuNoV genotype GII.4 is the dominant variant that causes most of the worldwide Norovirus outbreaks and the rapid genetic mutation rate emerged a new prevalent strain every few years [15]. Although there were several studies on the interaction between HuNoV and the host cells, understanding of HuNoV pathogenesis remains unclear due to its diverse genotypes [16,17]. To date, only the study on murine Norovirus (MuNoV) GV is able to demonstrate the life cycle of Norovirus [18-20]. The findings on MuNoV have become the model system to increase the understanding on another Norovirus gene expression and replication. Therefore the life cycle of Norovirus has been proposed based on studies on MuNoV.
Norovirus genome replication upon host cell entry
The succession of MuNoV to be propagated in vitro has led to various research in gaining the deeper insights of Norovirus biology. It has been suggested that noroviruses primarily interact with specific receptors lining the human intestinal epithelium prior to host cell entry. Nevertheless, HuNoV pathogenicity was proposed according to MuNoV propagation studied previously [18-22]. Foremost, the virus infectivity is initiated from the virus binding to a type of glycans known as histo-blood group antigens (HBGA) [23-25]. The HBGA are suggested to become the receptor or co-receptor binding site [26]. Several other studies have indicated that the lack of HBGA expression reduce the replication of Norovirus [27,28]. Notably, the Norovirus are proposed to enter the host intestine and released the RNA genome into the cytoplasm of permissive cells. Once uncoated, the VPg-RNA genome are thought to represent as the mRNA template for viral RNA translation. The VPg may initiate the translation of viral protein by performing as the cap substitute and engage with host cell translation initiation factor. The viral protease (Pro) cut the ORF 1 polyproteins during co- and post-translational phases prior to replication complex assembly via the interaction with amino acid protein (p48) and 3A-like protein (p22). The replication of virus genome begins from the negative-strand intermediate and RNA-dependant RNA polymerase (Pol) to produce the genomic and subgenomic RNA via de novo RNA synthesis. The genome product is further translated to form a new batch of genomic RNA or assemble into the major capsid protein (VP 1) before virus exits prior to other infections. The succession of MuNoV RNA replication was reported to cause lysis of macrophages, dendritic cell and B cell [29,30]. Contradicting results were obtained where no effects of HuNoV infection towards respective cells tested and the HuNoV RNA remained undetected or at least showed a modest increase, which indicated inefficient viral replication system. Nevertheless, there is still possibility that HuNoV cultivation can be successful considering the proven GV Norovirus replication and infinite culture system approaches tested.
Propagation of Human Norovirus (HuNoV)
In vitro cultivation of HuNoV
Various attempts have been made in cultivating human Norovirus (HuNoV) over the past two decades (Table 1). Based on the previous studies, the in vitro propagation of MuNoV on murine B cells showed the increasing MuNoV-RNA titre up to four-fold [29]. Following study reported a successful replication of HuNoV in the B cells indicated by the five-fold increase of the genome virus copies [31]. Nevertheless, a reliable cell culture system for HuNoV cultivation remains inconclusive imposing major challenges in understanding the virus life cycle. Furthermore, most of the approaches were unable to produce reproducible results hence limiting the study of the HuNoV biology due to limited source of virus and unreliable cell culture model system [32].
| Sources of HuNoV | Type of cell lines | Results | Ref. |
|---|---|---|---|
| GI and GII isolates from stool sample of gastroenteritis patient | Int 407 | Cytophatic effect (CPE) and RT-PCR positive for Norovirus | [3] |
| MuNoV-1 | Macrophages and dendritic cell | Both cell types showed MuNoV-1 RNA replication | [19] |
| MuNoV-1 and MuNoV-3 | M12 and WEHI-231 mouse B cell | Succession of viral infection and replication up to four-fold increase RNA titre. | [29] |
| GII.4 Sydney virus positive stool sample | B cells | Succession of virus replication with at least fivefold increase in genome copy number. | [31] |
| Virus-like particle MVA/T7 | HEK293T cells and BHK-21 | Replication of Norovirus genomic RNA expressed in mammalian cells | [32] |
| Calicivirus positive stool samples of gastroenteritis patients | Human intestinal (8), other human tissue (8), monkey kidney epithelia (4), other animal tissue (7) | All types of cells failed to demonstrate HuNoV replication | [33] |
| GII.4 isolates from positive stool sample | Caco-2 | Replication of viral RNA more than 2 Log10 | [34] |
| GI.1, GII.4 and GII.12 isolates from stool samples of patients | 3D culture of Int 407 and Caco-2 | Viral capsid protein VP1 located on the cells surface, no RNA viral replication occurs. | [35] |
| GI and GII isolates from stool sample of gastroenteritis patient | 3D culture of Int 407 and Caco-2 | Both cell types failed to demonstrate HuNoV replication | [36] |
| GII.4 isolates from positive stool sample | 3D culture of C2BBe1 | Replication of viral RNA more than 2 Log10 | [37] |
| Multiple GII.4 isolates | Human intestinal enteroids | Complete viral replication | [41] |
Table 1: List of various attempts for in vitro propagation of human Norovirus.
The search for efficient cultivation medium for HuNoV started when Duizer et al. [33] tested several cell lines including primary kidney cell lines, primary intestinal cell line and colon carcinoma cell line. However none of them were able to provide a reproducible in vitro cultivation system for HuNoV propagation most probably due to the inadequate specific factor such as secretor and binding protein from the host cell, the virus or both [33].
Previous study conducted by Straub et al. [34] also failed to demonstrate the propagation of HuNoV. The researchers suggested that innate immunity of host cell might limit the RNA replication. The similar results was obtained by Takanashi et al. [35] who found that HuNoV did infect the permissive cell, but did not replicate due to the possibility that its entry was blocked by the accumulation of virus inoculum as a result of microvilli beating. In addition, Papafragcou et al. [36] reported that the HuNoV were able to interact with host cell, however the replication of virus did not occur after the viral binding to an identified receptor, most likely due to host cells response that terminated the entry. Despite previous unsuccessful attempts, Jones et al. [31] made a ground breaking advance in 2015 whereby the researchers justified B cell as the cell culture system that supported the HuNoV replication. The finding from the study has provided a prospective cell culture model that is potentially reliable for HuNoV cultivation.
However, another limitation in developing an efficient HuNoV cultivation system is the reproducibility of the model. For instance, the model system developed by University of Michigans (Wobus Group) and Erasmus Medical Center (Koopmans Group) at University of Florida was unsuccessful when repeated by the Vinje research group and University of Florida (Karst Group) at the CDC. This was possibly due to the usage of FBS from a different source or limited virus stock isolates (decrease more than fivefold viral genome). Other parameters such as medium components, tissue culture plastic disposables used might also played important role in the efficiency of virus replication [31]. Despite the crucial obstacles in providing suitable culture system, another pivotal problem faced in the previous studies was the low level of viral replication. While using the unfiltered sample increase the possibility of ‘missing’ HuNoV RNA, various undetermined factors might influence the replication of this virus. The presence of unknown inhibitor might cause the contradicted correlation between the viral input and level of infection. In addition, the modest viral replication might be masked by the bulk viral input [31]. The same results were observed where a decrease in viral titer was reported due to unclear viral input [37]. Besides, the group suggested that possible viral clumping and independent sampling might be the cause of the said problem.
The succession of MuNoV to propagate on macrophages and dendritic cells was reported by Karst et al. [18] and Wobus et al. [19]. Based on the results, both the authors agreed on the possibility of the natural host specificity that facilitates the viral RNA replication. As the Norovirus was proposed to target the small intestine epithelium with blunting and shortening microvilli as the primary site of infection [35], the findings have emphasized the understanding on the Norovirus life cycle and prompt the other further research towards in vitro cultivation of HuNoV.
In 2008, Souza et al. [38] reported that the Norovirus interact with epithelial cells of animal model small intestine and HuNoV infection towards human volunteer biopsies exposed significant effect on small intestine. Subsequently, a study that focused on gastrointestinal epithelial was conducted by Straub et al. [37] and found that HuNoV binds to HBGA of Caco-2 cells monolayer. The results were aligned with the reports of presence of functional glycosylase enzyme prior to HBGA expression had been shown to modulate HuNoV infection [39].
The aims to make HuNoV cultivable may rely on the other tropism factor, such as the appearance of gut commensal bacteria. Recent study by Oka et al. [40] indicated that the gut bacteria did not support the HuNoV replication. However, no replication of viral RNA was reported after the infection even without the addition of the gut bacteria. Hence, the role played by the microfloras in assisting the HuNoV propagation remains unexplored in this study. Nonetheless finding of the study was in contrast with the previous finding by Jones et al. [31]. The author stated that the presence of commensal bacteria might promote the infectivity of HuNoV with the increase number of HBGA that were expressed on the bacterial cell’s surface. Furthermore, he indicate that the receptor and commensal bacteria cofactor and additional cell type/different character may be crucial for HuNoV infection which promote susceptible binding of virus prior to host cell entry. Interestingly though HuNoV was shown to infect human intestinal enteroids derived from stem cells without the assistance of bacteria or external HBGA [41].
Based on the references reviewed in this chapter, it has come to the consensus that the in vitro culture system with the B cells is deemed most suitable for the cultivation of HuNoV. However, currently efforts are ongoing towards the improvement of this culture system, as reviewed in the next chapter.
Advances of co-culture techniques as a strategy for human Norovirus cultivation succession
Co-culture system has been widely used to reenact the human in vivo-like model that allowed the study of cell to cell interaction. The establishment of gastrointestinal co-culture, M-like cells had provided several advantages such as study of the interaction between intestinal cell and human body immune response, cell invasion by pathogens, development of drug discovery and administration of oral vaccine. The M cells, also known as microfold, membranous or microvillous cells are defined as specialized epithelial cells found in follicleassociated epithelia (FAE) of the Peyer’s patches. The role of M cells includes the induction of immune responses within mucosa-associated lymphoid tissue (MALT) and transcytosis which a process of particles such as antigen and bacteria transportation from the apical side into lymphocytes pocketslike structure at the basolateral side. The M cells showed unique morphological structure with the presence of irregular brush border and reduced glycocalyx and microvilli at the apical surface, comprised approximately 10% of the epithelial cell within FAE [42]. Despite lack of M cells on human epithelial line, these specialized cells might be exploited by pathogens upon their host cell entry prior to host infection. Certain microorganisms and viruses manipulate the efficient transcytotic activity in facilitating its host invasion. There were several previous reports indicating that noroviruses might exploit the presence of M cells as the host entry site to breach human intestinal epithelium [29,43]. Nevertheless, the expression of specific surface protein receptors of M cells that might aid in particle-adhesion activity has also been reported previously. The finding of B1-integrin and glycoprotein 2 expressed by the M cells that acts as the surface receptor has made a presumptive indication of the infectivity assistance [42,44]. These data had gave a preliminary exposure on the role of surface protein expressed thus providing a promising knowledge on HuNoV pathogenesis via the M cells.
The relationship with M cells was also studied against murine Norovirus (MuNoV) [43]. The research was mainly focusing on either the virus infects and/or cross the intestinal barrier through M cell. It was observed that the MuNoV does not replicate in both monolayer and murine B cells, proposing that MuNoV overcame the epithelial intestinal barrier through M cell. Monolayer itself was also capable to trancytose particle with the appearance or absent of B cells. Apart from that, the addition of B cells does not increase the M cell formation, instead enhance the trancytosis by the secretion of cytokine and ETS transcription factor.
It has been hypothesized that HuNoV must breach and pass over the confluent epithelial monolayer co-culture system to reach B cells that underlying the basolateral compartment [31]. The authors indicated to simplify the culture techniques by having to use common medium and basal media. Nonetheless, modest level of replication culture system may contribute to a successful and reproducible HuNoV propagation system. In this study, they discover the cofactor for viral infection in which supported the findings by Gonzalez-Hernandez et al [43] while Hsieh et al. [45] reported that M cell maturity require secreted factor CD137. Hypothesis of the role of secretor was supported by Straub et al. [37] while Takanashi et al. [35] stated that secretors carry the FUT2 genes that was expressing the HBGA and thus vulnerable to HuNoV infection.
Concluding Remarks
To date, there is no efficient and reproducible cell culture system that is reliable for the human Norovirus (HuNoV). However, the new targeted M cells are seen as the promising approach in the search for a respective model system. In addition, several other recommendations can be considered such as the application of primary intestinal cell and human pluripotent stem cells. Furthermore, cells other than intestinal epithelial cell origin also can be used, as Norovirus are able to propagate in macrophages, dendritic and mesenchymal cell as described previously. Apart from that, gut commensal bacteria can be exploitied during the cultivation due to their possible significant role in facilitating HuNoV binding and host cell entry. The advancement of molecular biology field has led to the innovation of feasible approaches to improve the knowledge on the HuNoV pathogenesis. Despite the limited sources of HuNoV, the construction of virus-like particle (VLP) via DNA recombinant technology seems favorable to overcome the issue. The development of an effective in vitro system that is reproducible for the propagation of HuNoV is necessary for better understanding on the virus pathogenicity. A successful in vitro propagation system will lead to the development of HuNoV vaccine while sealing the fundamental gaps of HuNoV biology at the same time.
Acknowledgement
This study was financed by Fundamental Research Grant Scheme from Ministry of Education, Malaysia (600-RMI/ FRGS 5/367/2015) and was done at Universiti Teknologi MARA, Sungai Buloh, Malaysia.
References
- Hall AJ, Lopman BA, Payne DC, Patel MM, Gastañaduy PA, Vinjé J, Parashar UD. Norovirus disease in the United States. Emerg Infect Dis 2013; 19: 1198-1205.
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015; 12: 1001921.
- Straub TM, Bentrup KH, Orosz-Coghlan P, Dohnalkova A, Mayer BK, Bartholomew RA, Valdez CO, Bruckner-Lea CJ, Gerba CP, Abbaszadegan M, Nickerson CA. In vitro cell culture infectivity assay for Human Norovirus. Emerg Infect Dis 2007; 13: 396-403.
- Zahorsky J. Hyperemesis hiemis or the winter vomiting disease. Arch Pediat 1929; 46: 391-395.
- Kim JS, Kim HS, Hyun J, Kim HS, Song W. Molecular epidemiology of human Norovirus in Korea in 2013. Biomed Res Int 2015; 2015: 468304.
- Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology 1993; 195: 51-61.
- Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med 2009; 361: 1776-1785.
- Lay MK, Atmar RL, Guix S, Bharadwaj U, He H, Neill FH, Sastry KJ, Yao Q, Estes MK. Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans. Virol 2010; 406: 1-11.
- Nguyen GT, Phan K, Teng I, Pu J, Watanabe T. A systematic review and meta-analysis of the prevalence of norovirus in cases of gastroenteritis in developing countries. Medicine (Baltimore) 2017; 96: 8139.
- Debbink K, Lindesmith LC, Donaldson EF, Baric RS. Norovirus immunity and the great escape. PLoS Pathog 2012; 8: 1002921.
- Thorne LG, Goodfellow IG. Norovirus gene expression and replication. J Gen Virol 2014; 95: 278-291.
- Almand EA, Moore MD, Jaykus LA. Norovirus binding to ligands beyond histo-blood group antigens. Front Microbiol 2017; 8: 2549.
- Donaldson EF, Lindesmith LC, LoBue AD, Baric RS. Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol 2010; 8: 231-241.
- Kroneman A, Vega E, Vennema H, Vinjé J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol 2013; 158: 2059-2068.
- Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinjé J, Baric RS. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J Infect Dis 2013; 208: 1877-1887.
- Almand EA, Moore MD, Outlaw J, Jaykus LA. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS One 2017; 12: 0173124.
- Donaldson EF, Lindesmith LC, LoBue AD. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human population. Immunol Rev 2008; 225: 190-211.
- Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW 4th. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2003; 299: 1575-1578.
- Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2004; 2: 432.
- Lopez-Manriquez E, Vashist S, Urena L, Goodfellow I, Chavez P, Mora-Heredia JE, Cancio-Lonches C, Garrido E, Gutierrez-Escolano AL. Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1. J Virol 2013; 87: 11371-11387.
- Daugghenbaugh KF, Wobus CE, Hardy ME. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J 2006; 3.
- Furman LM, Maaty WS, Petersen LK, Ettayebi K, Hardy ME, Bothner B. Cysteine protease activation and apoptosis in murine norovirus infection. Virol J 2009; 139.
- Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 2007; 81: 5949-5957.
- Choi JM, Hutson AM, Estes MK, Prasad BV. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci USA 2008; 105: 9175-9180.
- Kubota T, Kumagai A, Ito H, Furukawa S, Someya Y, Takeda N, Ishii K, Wakita T, Narimatsu H, Shirato H. Structural basis for the recognition of Lewis antigens by genogroup I norovirus. J Virol 2012; 86: 11138-11150.
- Singh BK, Leuthold MM, Hansman GS. Structural constraints on human norovirus binding to histo-blood group antigens. mSphere 2016; 1: 00049-16.
- Herbst-Kralovetz MM, Radtke AL, Lay MK, Hjelm BE, Bolick AN, Sarker SS, Atmar RL, Kingsley DH, Arntzen CJ, Estes MK, Nickerson CA. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg Infect Dis 2013; 19: 431-437.
- Carmona-Vicente N, Allen DJ, Rodriguez-Diaz J, Iturriza-gomara M, Buesa J. Antibodies against Lewis antigens inhibit the binding of human norovirus GII.4 virus-like particles to saliva but not to intestinal Caco-2 cells. Virol J 2016; 13.
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014; 346: 755-759.
- Karst SM, Zhu S, Goodfellow IG. The molecular pathology of noroviruses. J Pathol 2015; 235: 206-216.
- Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, Freiden P, Graves CL, Koopmans M, Wallet SM, Tibbetts SA, Schultz-Cherry S, Wobus CE, Vinje J, Karst SM. Human norovirus culture in B cells. Nat Protoc 2015; 10: 1939-1947.
- Asanaka M, Atmar RL, Ruvolo V, Crawford SE, Neill FH, Estes MK. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc Natl Acad Sci USA 2005; 102: 10327-10332.
- Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. J Gen Virol 2004; 85: 79-87.
- Straub TM, Bartholomew RA, Valdez CO, Valentine NB, Dohnalkova A, Ozanich RM, Bruckner-Lea CJ, Call DR. Human norovirus infection of Caco-2 cells grown as a 3-dimensional tissue structure. J Water Health 2011; 9: 225-240.
- Takanashi S, Saif LJ, Hughes JH, Meulia T, Jung K, Scheuer KA, Wang Q. Failure of propagation of human norovirus in intestinal epithelial cells with microvilli grown in three-dimensional cultures. Arch Virol 2014; 159: 257-266.
- Papafragcou E, Hewitt J, Park GW, Greening G, Vinje J. Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PLoS One 2013; 8: 63485.
- Straub TM, Hutchison JR, Bartholomew RA, Valdez CO, Valentine NB, Dohnalkova A, Ozanich RM, Bruckner-Lea CJ. Defining cell culture conditions to improve human norovirus infectivity assays. Water Sci Technol 2013; 67: 863-868.
- Souza M, Avesado MSP, Jung K, Cheetham S, Saif LJ. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. J Virol 2008; 82: 1777-1786.
- Patel MM, Hall AJ, Vinje J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol 2009; 44: 1-8.
- Oka T, Stoltzfus GT, Zhu C, Jung K, Wang Q, Saif LJ. Attempts to grow human noroviruses, a sapovirus, and a bovine norovirus in vitro. PLoS One 2018; 13: 0178157.
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016; 353: 1387-1393.
- Mabbott NA, Donalson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveilance posts in the intestinal epithelium. Mucosal Immunol 2013; 6: 666-677.
- Gonzalez-Hernandez MB, Liu T, Payne HC, Stencel-Baerenwald J, Ikizler M, Yagita H, Dermody TS, Williams IR, Wobus CE. Efficient norovirus and reovirus replication in the mouse intestine requires microfold (M) cells. J Virol 2014; 88: 6934-6943.
- Ohno H. Intestinal M cells. J Biochem 2016; 159: 151-160.
- Hsieh EH, Fernandez X, Wang J, Hamer M, Calvillo S, Croft M, Kwon BS, Lo DD. CD137 is required for M cell functional maturation but not lineage commitment. Am J Pathol 2010; 177: 666-676.