ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 11
Abnormality detection using weighed particle swarm optimization and smooth support vector machine
Latchoumi TP1 and Latha Parthiban2*
1Research Scholar, Department of Computer Science and Engineering, Sathyabama University, Assistant Professor, Vignan’s University, Vadlamudi, Andra Pradesh, India
2Department of Computer Science, Pondicherry University CC, Pondicherry, Tamil Nadu, India
- *Corresponding Author:
- Latha Parthiban
Department of Computer Science, Pondicherry University CC
Pondicherry, Tamil Nadu, India
Accepted date: March 14, 2017
In this paper, a new hybrid classification approach, which uses Weighted-Particle Swarm Optimization (WPSO) for data clustering in sequence with Smooth Support Vector Machine (SSVM) for classification is proposed. The performance of WPSO clustering is compared with K means and fuzzy methods using intercluster, intracluster and validity index. The accuracy of proposed WPSO-SSVM classification methodology are 83.76% for liver disorder, 98.42% for WBCD, 95.21% for mammographic mass data which are better than in existing literature.
Keywords
Smooth support vector machine (SSVM), Particle swarm optimization (PSO), Clustering, Classification.
Introduction
Medical data mining has a great potential for exploring hidden patterns and extracting useful information for decision support [1]. Benefits of introducing machine learning into medical analysis are to increase diagnostic accuracy, reduce costs and human resources [2]. Case based reasoning [3] process is an approach for developing knowledge-based medical decision support system which solves new problems based on the solutions of similar past problems.
Materials and Methods
Assume a medical library with each case in the library as index of corresponding features (e1, e2, ..., eN) having an associated action, with collection of features Fj (j=1…. n) representing the cases and variable V denoting the action. The ith case ej in the library can be represented as an n+1-dimensional vector, i.e. ei=(xi1, xi2, ......, xin, yi). Where xij corresponds to the value of feature Fj (j=1... n) and yi corresponds to the action (i=1... n). If for each j (1 ≤ j ≤ n) a weight wj (wj (0, 1)) has been assigned to the jth feature to indicate the importance of the feature, then for any pair of cases ep and eq in the library, a weighted distance metric dpq(w) is defined as:
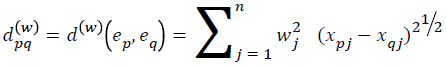
Where xpj is the pth case with jth feature and xqj is the qth case with jth feature. Using the weighted distance a similarity measure SMpq(w) is calculated usingSMpq(w)=1/(1+αdpq(w))
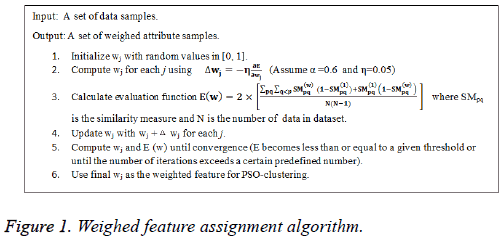
Where α is a positive parameter. The weighted feature assignment algorithm is presented in Figure 1.
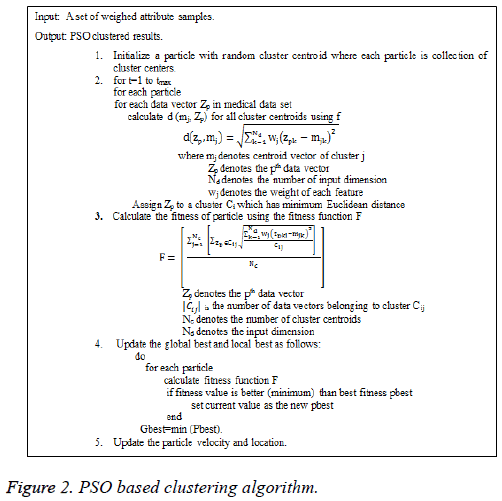
PSO is a population-based search algorithm and each particle is associated with a velocity and its algorithm is presented in Figure 2.
A nonlinear version of the SSVM [4] is used for classification of datasets after clustering.
Results
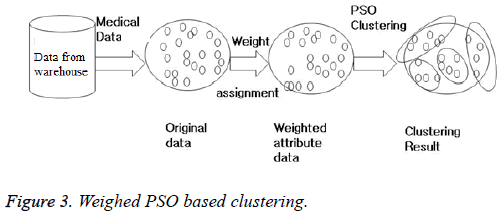
The WBCD, mammographic mass and liver disorder dataset are obtained from UCI machine learning repository [5]. Weighed PSO clustering is applied on the datasets (Figure 3) and compared with K-means and FCM in terms of intercluster, intra cluster and validity index as shown in Table 1. The inter cluster distance of any two cluster should be high which is best for PSO as seen in Table 1. Intra cluster means the compactness of a cluster and its value should be least as possible and is again best for PSO.
| Measures | FCM | K-means | PSO | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WBCD | Liver disorder | Mammographic mass | WBCD | Liver disorder | Mammographic mass | WBCD | Liver disorder | Mammographic mass | |
| Inter Cluster | 708.56 | 87.4948 | 24.43 | 713.944 | 109.817 | 23.91 | 941.771 | 172.3005 | 237.639 |
| Intra Cluster | NA | NA | NA | 11.4572 | 2.6474 | 0.3942 | 0.292068 | 0.0846 | 0.004 |
| Validity Index | NA | NA | NA | 0.016047 | 0.0241 | 0.0164 | 0.00031 | 0.0049 | 0.0001 |
Table 1: Comparison of inter, intra and validity index with FCM, K-means and PSO for breast cancer (WBCD) and Liver disorder dataset [6].
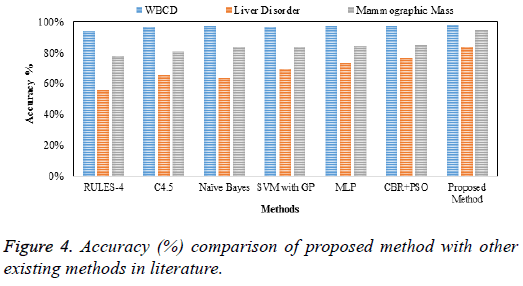
The clustered output is classified using SSVM using fivefold cross validation in which randomly split database is averaged to provide the best indication of true classification performance and the performance comparison of datasets is presented in Table 2 and accuracy is shown in Figure 4.
| Methods | WBCD | Liver Disorder | Mammographic Mass | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |
| RULES-4 | 94.74% | 96.43% | 92.56% | 55.90% | 56.78% | 54.57% | 78.13% | 79.55% | 75.67% |
| C4.5 | 96.80% | 97.12% | 94.54% | 65.59% | 66.78% | 64.85% | 81.13% | 84.54% | 79.56% |
| Naive Bayes | 97.36% | 98.53% | 96.23% | 63.39% | 66.45% | 61.23% | 83.43% | 86.64% | 82.36% |
| SVM with GP | 96.70% | 98.40% | 94.97% | 69.70% | 71.67% | 65.67% | 83.66% | 85.54% | 81.14% |
| MLP | 97.20% | 98.57% | 96.25% | 73.05% | 74.57% | 72.46% | 84.79% | 87.64% | 82.45% |
| CBR+PSO | 97.41% | 98.53% | 96.45% | 76.81% | 77.67% | 73.68% | 85.29% | 87.64% | 83.44% |
| Proposed method | 98.42% | 99.38% | 97.35% | 83.16% | 86.16% | 77.17% | 95.21% | 97.57% | 93.45% |
Table 2: Performance comparison of datasets.
Conclusions
This paper proposes a new WPSO-SSVM technique to improve the classification accuracy of medical datasets and the obtained results are found to outperform all the present stateof- art classifiers existing in literature. The future work will be to test the proposed technique in other benchmark datasets to prove the robustness of the proposed algorithm.
References
- Pei CC, Jyun JL, Chen HL. An attribute weight assignment and particle swarm optimization algorithm for medical database classifications. Comp Met Prog Biomed 2011; 1-11.
- Bojarczuk HSL, Freitas AA. Genetic programming for knowledge discovery in chest-pain diagnosis: exploring a promising data mining approach. IEEE Eng Med BiolMagaz 2000; 19: 38- 44.
- Klaus DA. Case-based reasoning for medical decision support tasks: the inreca approach. Artificial Intel Med 1998; 12: 25-41.
- Yuh-Jye L,Mangasarian. Smooth support vector machine. Comp OptimizAppl2001; 20: 5-22.
- Data Mining Institute, University of Wisconsin, Technical Report 99-03.
- https://archive.ics.uci.edu/ml/datasets.html (Liver disorders, Mammographic mass and Wisconsin Breast Cancer (Original)).