ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 4
Alternative versus gold standard method for the typing of vancomycinresistant Enterococcus faecium: semi-automated rep-PCR system
1Ankara Numune Training and Research Hospital, Medical Microbiology, Ankara, Turkey
2Public Health Agency of Turkey, National Molecular Microbiology Reference Centers Laboratory, Ankara, Turkey
3Departmen of Medical Microbiology, Faculty of Medicine Yildirim Beyazit University, Ankara, Turkey
4Adana Numune Training and Research Hospital, Medical Microbiology, Adana, Turkey
- *Corresponding Author:
- Sumeyra Savas Acar
Public Health Agency of Turkey
National Molecular Microbiology
Reference Centers, Laboratory, Turkey
Accepted on August 8, 2016
Vancomycin-Resistant Enterococcus faecium (VRE) are significant nosocomial pathogens with limited treatment options. Vancomycin-Resistant Enterococcus faecium (VRE) has become possible transfer of vancomycin resistance to more virulent pathogens. The current study examined the detection of inhospital distribution and the spread of Vancomycin-Resistant Enterococcus faecium (VRE), in addition to the comparison of the availability of the methods of Pulsed Field Gel Electrophoresis (PFGE) and the Rep-PCR DiversiLab method in the detection of the clonal association between Vancomycin-Resistant Enterococcus faecium (VRE) isolates. Twenty-two humans and twenty-two environmentally originated isolates isolated from different wards were included in the study. A total of four clones were identified by the Rep-PCR DiversiLab during the evaluation after the finger analysis. Six isolates were found to belong none of the clones. On the other hand, Pulsed Field Gel Electrophoresis (PFGE) typing yielded five Pulsed Field Gel Electrophoresis (PFGE) including a total of 47 clonally related isolates and one unique isolate. As a result of one-to-one comparisons of the PFGE DNA patterns of a total of 48 species. The fact that the isolates were isolated from different clinics and samples suggested that the majority of Vancomycin-Resistant Enterococcus faecium (VRE) isolates in the hospital environment spread through cross contamination. The Pulsed Field Gel Electrophoresis (PFGE) method had greater reliability and differentiating capacity according to the comparison of both methods using epidemiological data.
Keywords
Vancomycin-resistant Enterococcus faecium (VRE), Pulsed field gel electrophoresis, Rep-PCR.
Introduction
Enterococci, especially the Vancomycin Resistant Enterococci (VRE) with limited options for treatment, are significant nosocomial pathogens. Enterococci, although they have a low virulence, may settle in the hospitals and cause serious infections since they demonstrate multi-drug resistance [1,2]. Urinary tract infections, bacteraemia, and endocarditis are frequently caused by these bacteria in hospital [3]. Vancomycin resistant enterococci in patients are associated with serious outcomes such as increased mortality, morbidity, and medical costs [4]. The determination of the multi-antibiotic resistance of enterococci and appearance of evidence of exogenous acquisition of enteroccal infections has increased the necessity of epidemiological studies and identification of the species [5]. Classical typing methods such as bacteriocin (enterococci) typing, phage typing, biochemical reaction profiles, antimicrobial resistance patterns, and serologic methods have been used in the evaluation of hospital epidemiology. However,classical phenotypical methods generally could not adequately differentiate enterococci. Molecular typing plays an important role in the evaluation of resistant enterococci, and the prevention and control of their spreads [5,6]. Pulsed Field Gel Electrophoresis (PFGE) has been proven to be useful for high discriminatory power, so that Pulsed Field Gel Electrophoresis (PFGE) is considered the gold standard reference method for molecular typing of enterococci. However, the main limitation of Pulsed Field Gel Electrophoresis (PFGE) include its being time consuming, requiring experienced personnel, is technically variable, and is expensive [7-9]. DiversiLab System (DL) (bioMerieux, Marcy Etoile, France) was introduced a semi-automated rep-PCR system. It has more advantages than rep-PCR, such as higher reproducibility, standardization, and archiving fingerprinting patterns and reporting [3,10-12]. The commercialized DiversiLab system (BioMerieux, Marcy I’Etoile, France) is a semi-automated rep-PCR technique [7]. This system has some noticeable advantages, such as simplistic data elaboration, production of results within one day, fingerprinting patterns, higher reproducibility, and visualization of the data. Additionally, strains, are compared over time to understand the charting the epidemiological of isolates with this system [7,8,11-15]. We investigated the two methods to distinguish the clonal relationship among epidemiologically well-defined isolates of vancomycin resistant enterococci. Furthermore, the technical properties of the DiversiLab System were compared with Pulsed Field Gel Electrophoresis (PFGE). The determination of the epidemiological and genotypic associations of the isolates would provide important benefits in the prevention of or decrease in the rate of hospital infections. It is expected that this knowledge will support clonal investigations of vancomycin resistant enterococci around the world.
Materials and Methods
Bacterial isolates
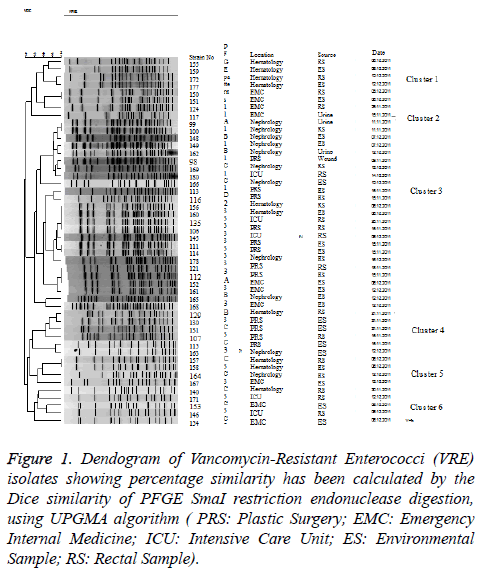
Forty-eight Vancomycin-Resistant Enterococci (VRE) isolated from rectal swab samples obtained from the patients hospitalized in various clinics in the context of a surveillance program conducted by the Infection Control Committee of the Numune Education and Research Hospital, Ministry of Health, and environment cultures and clinical materials between November 9, 2011 and December 14, 2011 were included in this study. Eighteen (37.5%) of the samples were rectal swab samples and 26 (54.2%) were obtained from the environment cultures taken from the rooms of patients colonized/infected with Vancomycin-Resistant Enterococcus faecium (VRE). Seventeen of the cases (65.4%) were isolated from the bed rails, four (15.4%) from the whatnots, two (7.7%) from the closets, and one each (3.9%) from the bedside table, dressing cart, and mobile ultrasonography equipment, respectively. Detailed information about the strains is presented in Table 1.
| Services | Locations | Sources | Strain No |
|---|---|---|---|
| PRS | Couch | ES | 111, 112, 113, 114, 115, 130 |
| Whatnot | ES | 131 | |
| Sickhuman | RS | 106, 121, 107 | |
| Wound | 98 | ||
| Haematology | Couch | ES | 158, 159, 177 |
| Deception | ES | 160 | |
| Sickhuman | RS | 155, 156, 129, 157, 140, 172 | |
| Nephrology | Couch | ES | 149, 178, 164, 165 |
| Whatnot | ES | 166 | |
| Deception | ES | 148 | |
| Sickhuman | RS | 100, 169 | |
| Urine | 99, 162 | ||
| EMC | Couch | ES | 151, 152, 153, 154 |
| Whatnot | ES | 167, 168 | |
| Sickhuman | RS | 150, 124 | |
| Urine | 117 | ||
| ICU | Sickhuman | RS | 171, 180, 135, 145, 146 |
Table 1. Basic epidemiological data of the 48 distribution of Vancomycin-Resistant Enterococcus faecium (VRE) isolates (PRS: Plastic Surgery; EMC: Emergency Internal Medicine; ICU: Intensive Care Unit; ES: Environmental Sample; RS: Rectal Sample).
Sample collection and cultivation
The cultivation of rectal swabs and environmental samples obtained by sterile swabs were performed on an enterococcus agar plate (Oxoid, United Kingdom). They were incubated for a maximum of 72 hours at 37˚C in an aerobic medium. Environmental samples were incubated in Vancomycin- Resistant Enterococci (VRE) broth agar (Oxoid, United Kingdom) for 24 hours at 37˚C prior to cultivation in enterococcus agar plate. Colonies growing black in color in the enterococcus agar plates and suspicious colonies of clinical samples observed in sheep blood agar were evaluated using conventional methods and defined as Enterococcus spp. Strain definitions and antibiotic sensitivity were performed using a Vitek-2 automated system according to the directions of the manufacturer.
Pulsed-field gel electrophoresis (PFGE)
Whole cellular DNA for Pulsed Field Gel Electrophoresis (PFGE) method was employed in accordance with Durmaz et al. [16]. Agarose plugs were prepared and then digested with 10 U/μL SmaI (Fermentas Life Sciences, St. Leon-Rot, Germany) restriction enzyme. Electrophoresis was performed with 1% SeaKem Gold agarose gel in 0.5 X Tris-Boric Acid- EDTA buffer using the CHEF DRII system (Bio-Rad, Hercules, USA). The total run time was 20 h; switch time was 5.3 to 34.9 s at 6 V/cm, 140˚C. The gels were stained with ethidium bromide (2 mg/mL, Sigma) for 15 min and washed thrice with distilled water for 15 minutes and visualized using a UV transilluminator. The restriction patterns were compared by using the Bionumerics version 6.01 software with the Dice coefficient with 1.5% band tolerance and 1% optimization and the Unweighted Pair Group Method with Arithmetic Averages (UPGMA). The clonal relationship among isolates was evaluated using the criteria of Tenover et al. [17].
Rep PCR using DiversiLab system
The genetic relatedness of the Vancomycin-Resistant Enterococci (VRE) isolates was determined by Rep-PCR typing as previously described [18]. Briefly, Vancomycin- Resistant Enterococci (VRE) organisms were cultured overnight and the DNA was extracted using an Ultra-Clean microbial DNA isolation kit (MO BIO Laboratories Inc., California, ABD) DiversiLab Enterococcus Rep-PCR was performed using the DiversiLab Enterococcus kit (Biomerieux SA, Marcy l’ Etoile, France). The Rep-PCR products were analysed by the DiversiLab system and software (Bacterial Bar Codes, Spectral Genomics, Houston, TX). The DNA fingerprint patterns were presented as virtual electropherograms and scatter plot analysis. The analysis was performed with DiversiLab software version 3.4 using the Pearson correlation coefficients to determine genetic similarities to create dendrograms. Samples were classified into the same Rep-PCR group if the similarity was >97% [18].
Results
Differentiation of Vancomycin-Resistant Enterococcus faecium (VRE) isolates by Pulsed Field Gel Electrophoresis
The 48 Vancomycin-Resistant Enterococci (VRE) isolates were separated into six major SmaI PFGE types. These six different Pulsed Field Gel Electrophoresis (PFGE) types were separated into five groups and one unique profile based on their Pulsed Field Gel Electrophoresis (PFGE) bands and their percentages. The overall level of genetic relatedness among the six Pulsed Field Gel Electrophoresis (PFGE) types was between 80%-85% similarity. As a result of the Pulsed Field Gel Electrophoresis (PFGE) study, seven of 48 Vancomycin- Resistant Enterococci (VRE) isolates were determined to belong to Pulsed Field Gel Electrophoresis (PFGE) Group 1, one was determined as unique, 25 to Group 2, six to Group 3, four to Group 4, and five belonged to Group 5. Among the isolates, Pulsed Field Gel Electrophoresis (PFGE) Group 1 included five variants and Group 2, Group 3, Group 4, and Group 5 had 6, 4, 3, and 4 different variants, respectively. The Pulsed Field Gel Electrophoresis dendrogram image was shown in Figure 1.
Figure 1: Dendogram of Vancomycin-Resistant Enterococci (VRE)isolates showing percentage similarity has been calculated by the Dice similarity of PFGE SmaI restriction endonuclease digestion,using UPGMA algorithm ( PRS: Plastic Surgery; EMC: Emergency Internal Medicine; ICU: Intensive Care Unit; ES: Environmental Sample; RS: Rectal Sample).
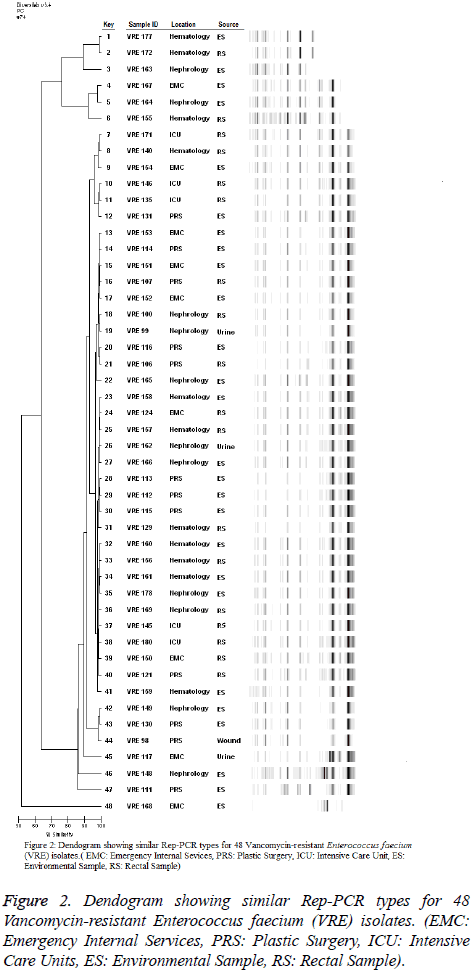
Differentiation of Vancomycin-Resistant Enterococcus faecium (VRE) isolates by Rep-PCR
A total of four clones were identified by Rep-PCR during the evaluation performed after fingerprint analysis to determine the genetic associations of Vancomycin-Resistant Enterococci (VRE) isolates. Two, two, 35, and three Vancomycin-Resistant Enterococci (VRE) strains were present in the A, B, C, and D clones, respectively Figure 2. The similarity of the two isolated in the A clone was 99.2% and these two isolates were obtained from the rectal swab of a patient admitted to the haematology clinic and from the bed rail of the same patient. One of the isolates in the B clone was isolated from the whatnot in ADS and the other from the environmental culture sample taken from the bed rail in the nephrology clinic on the same day; their rate of similarity was 97.9%. The C clone with the greatest amount of isolates (35/48; 72.9%) was isolated from each of the wards in which the samples were obtained for the study. This clone was isolated in all (5/5), 75% (3/12 rectal swab, 6/12 environment), 72.73% (4/11 rectal swab, 4/11 environment), 66.67% (2/9 rectal swab, 4/9 environment), and 63.64% (2/11 urine; 2/11 rectal swab, 3/11 environment) of the samples sent from the ICU, plastic surgery ward, haematology ward, ADS, and nephrology ward, respectively. The C clone sample was obtained between November 11 and December 16, 2011. There were three isolates in clone D, one from a wound sample of a patient, one from an environment culture, and one from an environment culture in nephrology ward. In addition, six isolates (8.2%) including more than two bands and at the same time having a similarity of less than 95% with other strains were found to belong to none of the clones. Two of them were isolated from ADS (1/6 urine; 1/6 environment), two from nephrology environment cultures, one from a PCS environment culture, and one from a rectal swab culture of a patient admitted to the haematology ward.
Discussion
Vancomycin resistance has been reported with gradually increasing frequency around the world since its first report from United Kingdom in 1986 and subsequently in 1987 from the United States. On the other hand, the first vancomycin resistant Escherichia faecium species was detected in Antalya in 1998 and subsequent reports from other regions followed this [19-21].
Isolates have to be identified at the level of strains and features of resistance should be determined to detect the epidemiological characteristics of the microorganisms, followed by the determination of relations between the isolates using molecular epidemiological methods.
Identification of the source and route of dissemination of Vancomycin-Resistant Enterococci (VRE) is helpful for controlling outbreaks [7].
Currently, although various molecular epidemiological methods have been tested in the detection of epidemiological characteristics of Vancomycin-Resistant Enterococcus faecium (VRE) outbreaks in particular, the Pulsed Field Gel Electrophoresis (PFGE) method is accepted as the “gold standard” due to its reproducibility, high rate of differentiation, and quantifiable results. This method is extremely beneficial in defining the outbreak strain, amount of distribution and source in hospital infection outbreaks developing by bacteria [22].
In this study, the performance of automated Rep-PCR was compared with Pulsed Field Gel Electrophoresis (PFGE) for 48 isolates collected a hospital from different locations and sources. When the results of the two methods were evaluated, a dominant cluster was observed (Pulsed Field Gel Electrophoresis (PFGE) Group 2, Rep PCR Group C). However, although the strains obtained from items and patient samples related with each other were observed to be independent samples by the DiversiLab system, the results of the Pulsed Field Gel Electrophoresis (PFGE) method were found to be consistent with the epidemiological data. In addition, large DL types were generally subdivided by Pulsed Field Gel Electrophoresis (PFGE) into several different types [8]. Therefore, the DiversiLab system had less discriminant power, creating larger clusters and clustering of isolates. Nevertheless, the DiversiLab system can be used in in-hospital outbreaks when obtaining rapid results and precautionary measures are needed. This is due to the fact that Rep-PCR using DiversiLab is semi-automated, microfluidic separation of fragments, a complete microbial typing analysis that includes rapid results in approximately four hours, versus 2-3 days for Pulsed Field Gel Electrophoresis (PFGE) [9,23]. According to Brolund et al., Lue et al. and Pitout et al., isolates classified as indistinguishable by DL therefore required confirmation with a method of higher resolution to avoid false reporting of probable epidemiological relatedness [8,24,25]. Pounder et al. and Choung et al. agreed that DiversiLab is easy to use and provides rapid results, but they generally agreed in a lack of discriminatory power compared to Pulsed Field Gel Electrophoresis (PFGE), as did the authors of the current study [11,26]. In contrast to the results of ours and many other studies [11,25,26]. Bourdon et al. reported that Rep-PCR appeared to be more discriminative than Pulsed Field Gel Electrophoresis (PFGE) [9]. Epidemiological data is very important for molecular typing studies. However, there were limitations, the lack of epidemiological data, to the previously published studies [8,11]. In our study, epidemiological data shows better correlation with the Pulsed Field Gel Electrophoresis (PFGE) results than DiversiLab.
In conclusion, the Vancomycin-Resistant Enterococci (VRE) agent in the hospital was considered to originate from a common Pulsed Field Gel Electrophoresis (PFGE) group (Group 2) dominating the hospital. However, the fact that this strain Pulsed Field Gel Electrophoresis (PFGE) group has different pulsotypes suggests that the strains in this major group, which have long been present in the hospital, have generated a genetic differentiation. In addition, infections due to minor pulsotypes have also been identified. The determination of resistance genotypes and the demonstration of clonal associations are important in the follow-up and prevention of infections due to resistant microorganisms. This study, which differs from the others, is important since it compares the Pulsed Field Gel Electrophoresis (PFGE) group and DiversiLab systems using detailed epidemiological data for Vancomycin-Resistant Enterococci (VRE) isolates. We conclude that the DiversiLab was shown to be technically simpler, have an international database to allow the comparison of isolates, and adhere to protocols and more easily learned than Pulsed Field Gel Electrophoresis. However, DiversiLab has a lower discriminatory power than that of Pulsed Field Gel Electrophoresis.
Acknowledgments
This study was presented at the 8th National Molecular and Microbiology Congress, Ankara, June 4-7, 2014.
Conflict of Interest
Conflict of Interest
References
- Cetinkaya Y. Resistant problems of enterococcus. Newly and one again to come to order infections. Ankara Sci Med Publ 2004; 10-16.
- Murray BE. The life and times of the Enterococcus. ClinMicrobiol Rev 1990; 3: 46-65.
- Shokoohizadeh L, Mobarez AM, Zali MR, Ranjbar R, Alebouyeh M, Sakinc T, Ali L. High frequency distribution of heterogeneous Vancomycin Resistant Enterococcousfaecium (VREfm) in Iranian hospitals. DiagnPathol 2013; 8: 163.
- Gordoncillo MJ, Donabedian S, Bartlett PC, Perri M, Zervos M, Kirkwood R, Febvay C. Isolation and characterization of Vancomycin-Resistant Enterococcus faecium from swine in Michigan, USA. Zoon Pub H 2013; 60: 319-326.
- Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH. Enterococcus. ClinMicrobiol 2009; 30: 430-442.
- Facklam RR, Sahm DF. Enterococcus. Manu Microbiol (6th edn.) 1995; 308-314.
- Bourdon N, Lemire A, Fines-Guyon M, Auzou M, Perichon B, Courvalin P, Cattoir V, Leclercq R. Comparison of four methods, including semi-automated rep-PCR, for the typing of vancomycin-resistant Enterococcus faecium. J Microbiol Methods 2011; 84: 74-80.
- Brolund A, Haeggman S, Edquist J, Gezelius L, Liljequist BO, Wisell KT. The DiversiLab system versus pulsed-field gel electrophoresis: Characterization of extended spectrum ß-lactamase producing Escherichia coli and Klebsiellapneumoniae. J Microbiolo Methods 2010; 83: 224-230.
- Kilic A, Bedir O, Kocak N, Levent B, Eyigun, CP. Analysis of an outbreak of Salmonella Enteritidis by repetitive-sequence-based PCR and pulsed field gel electrophoresis. Int Med 2010; 49: 31-36.
- Fontana C, Favaro M, Minelli S, Bossa MC, Testore GP, Leonardis F, Natoli S, Favalli C. Acinetobacterbaumannii in intensive care unit: a novel system to study clonal relationship among the isolates. BMC Infect Dis 2008; 8: 79.
- Pounder JI, Shutt CK, Schaecher BJ, Woods GL. Clinical evaluation of repetitive sequence-based polymerase chain reaction using the Diversi-Lab System for strain typing of vancomycin-resistant enterococci. DiagnMicrobiol Infect Dis 2006; 54: 183-187.
- Higgins PG, Hujer AM, Hujer KM, Bonomo RA, Seifert H. Interlaboratory reproducibility of DiversiLab rep-PCR typing and clustering of Acinetobacterbaumannii isolates. J Med Microbiol 2012; 61: 137-141.
- Voets GM, Leverstein-van Hall MA, Kolbe-Busch S, Van der Zanden A, Church D, Kaase M, Grisold A, Upton M, Cloutman-Green E, Conton R, Friedrich AW, Fluit AC. International Multicenter Evaluation of the DiversiLab Bacterial Typing System for Escherichia coli and Klebsiella spp. J ClinMicrobiol2013; 51, 12: 3944-3949.
- Pournaras S, Protonotariou E, Voulgari E, Kristo I, Dimitroulia E, Vitti D, Tsalidou M, Maniatis AN, Tsakris A, Sofianou D. Clonal spread of KPC-2 carbapenemase- producing Klebsiellapneumoniae strains in Greece. J AntimicrobChemother2009; 64: 348-352.
- Navon- Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y, Israeli KPC. First report on a hyperepidemic clone of KPC-3- producing Klebsiellapneumoniae in Israel genetically related on a strain causing outbreaks in the United States. Antimicrob Agents Che-mother 2009; 53: 818-820.
- Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y, Aktas E, Gursoy NC, Caliskan A. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacterbaumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis 2009; 62: 372-377.
- Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: A review for healthcare epidemiologists. Infect Control HospEpidemiol1997; 18: 426-439.
- Healy M, Huong J, Bittner T, Lising M, Frye S. Microbial DNA typing by automated repetitive-sequence-based PCR. J ClinMicrobiol 2005; 43: 199-207.
- Arslan U, Demir E, Oryasin E, Turk- Dagi H, Tuncer I, Findik D, Bozdogan B. MLST types of Vancomycin-Resistant Enterococcus faecium strains isolated from blood culture. MikrobiyolBul 2013; 47: 432-441.
- Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. ClinMicrobiol Rev 2000; 13: 686-707.
- Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinssom KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadow E, Simonsen, GS, Top J, VuopioVarkila J, Willems W, Woodford N. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill 2008; 13: 19046.
- Akcimen B. The genotyping of vancomycin resistant enterococci isolated nosocomial infections with pulsed field gel electrophoresis. CukurovaUni Med MicrobiolTurkey 2010.
- Ben-Darif E, Pinna E, Threlfall EJ, Bolton FJ. Comparison semi-automated rep-PCR system and multilocus sequence typing for differentiation of Salmonella enterica isolates. J Microbiol Methods 2010; 81: 11-16.
- Lau SH, Cheesborough J, Kaufmann ME, Woodford N, Dodgson AR, Dodgson KJ, Bolton EJ, Fox AJ, Upton M. Rapid identification of uropathogenic Escherichia coli of the O25:H4-ST131 clonal lineage using the DiversiLab repetitive-sequence based PCR system. ClinMicrobiol Infect 2010; 16: 232-237.
- Pitout JD, Campbell L, Church DL, Wang PW, Guttman DS, Gregson DB. Using a commercial DiversiLabsemiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J ClinMicrobiol2009; 47: 1212-1215.
- Chuang YC, Wang JT, Chen ML, Chen YC. Comparison of an automated repetitive-sequence-based PCR microbial typing system with pulsed- field gel electrophoresis for molecular typing of vancomycin-resistant Enterococcus faecium. J ClinMicrobiol 2010; 48: 2897-2901.