ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 1
ASP178ASNprion mutation causing transmissible spongiform encephalopathy under in-silico assessment using molecular dynamics simulation tool
1Department of Infectious diseasesHubei xiaogan central hospital infection, Hubei, PR China
2Department of Neurosurgery, Hubei xiaogan central hospital infection, Hubei, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Wei Dong
Department of Neurosurgery
Hubei xiaogan central hospital infection
Hubei
PR China
Accepted on 6 June, 2016
Transmissible spongiform encephalopathy’s caused by infectious agents is characterized as genetic or sporadic disorder. ASP178ASN (D178N) mutation as a genetic factor is well documented. The computational method was employed to look into the changes at atomic level, where disease associated prediction was followed by 50 nano second (ns) simulation under GROMOS96 43a1 force field at 300 K temperature. All the four servers used in this study predicted the D178N mutation to be highly deleterious. The simulations showed a considerable change in β-Sheet leading to change in intermolecular interactions and conformational rearrangements. These factors possibly contribute to pathogenic nature of D178N mutant prion.
Keywords
Periodontitis, hCAP, Mutation, Molecular dynamics simulation.
Introduction
Prion protein (PrP) is 253 aa long protein encoded by short (p) arm of chromosome 20 and is associated with human prion (HuPrP) diseases. The HuPrP diseases also known as Transmissible Spongiform Encephalopathy (TSE) has infectious, genetic and sporadic nature and is categorized into nine different disease variants, these diseases are attributed to the change of cellular PrP (PrPC) to its misfolded form known as scrapie PrP (PrPsc). The PrPsc is known to have more beta sheet and lesser alpha helix content than PrPC. PrPsc form aggregates that accumulate in the brain and ultimately leading to the pathogenesis of HuPrP diseases [1-3]. Five of the nine disease variants of TSE have genetic origin, where a mutation in PrP is playing a role in the etiology of TSE [2,4-8]. Asp178Asn (D178N) mutation mediated by polymorphism at residue 129 in PrP gene is a predominate factor in the etiology of TSE [9-11].
The PrPsc created by this mutation lead to TSE which is in accordance with “protein-only” hypothesis and its transmission does not require genome and propagates by binding of PrPsc with cellular prion protein and catalysing its conversion to the PrPSc state [12,13]. The molecular etiology of this transformation is poorly understood. However biophysical approaches are being employed in plethora to understand this transformation [14]. In this study we are studying the effect of D178N mutation on the transition of PrPC to PrPsc using atomic simulation. The Atomic insight into the protein architecture has been used to reveal the effect of the mutations [15,16] (Table 1).
| Method | Algorithm | Based on | Criteria | website | Damaging/Disease |
|---|---|---|---|---|---|
| SIFT | Alignment scores | Sequence alignment | 0-0.05 (Intolerant) | http://sift.jcvi.org/www/SIFT_enst_submit.html | YES |
| Phd-SNP | support vector machine | Sequence and profile information | Disease probability (if P>0.5 mutation is predicted Disease) | http://snps.path.uab.edu/phd-snp/phd-snp.html | YES |
| MutPred | random forest | SIFT and Gain/loss of functions | Scores with g>0.5 and p<0.05 are referred to as actionable hypotheses. | http://mutpred.mutdb.org/ | YES |
| Polyphen-2 | Bayesian classification | Physical and comparative considerations | Score ≥ 0.5 | http://genetics.bwh.harvard.edu/pph2/ | YES |
Table 1. D178N mutation deleterious prediction using four severs.
Material and Methods
Protein preparation
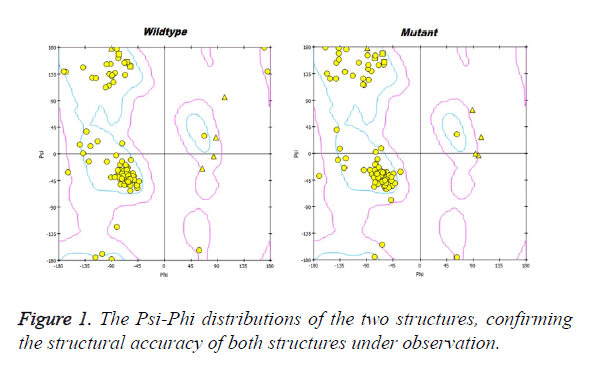
The prion protein crystallographic structure was retrieved from PDB repository (www.pdb.org), PDB ID: 3HAK [17]. The structure was energy minimized using SPDB viewer [18], the structure was used as wild-type (WT) and the mutation D178N was inserted and then the structure was energy minimized and was used as mutant (MU) in the study. The psi-phi distributions were studied before selecting the structures for further study.
Mutation damage prediction
Four in-silico servers were selected to check the deleterious effect of D178N on protein structure and function. Polymorphism effect prediction servers available online: SIFT [19], Polyphen-2 [20], PhD-SNP [21] and MutPred [22] were used.
Molecular dynamics simulation
Gromacs 4.5.6 package [23] was employed for molecular dynamics simulations (MDS) of WT and MU prion structures for 50 nano second (ns) under GROMOS96 43a1 force field in the NPT ensemble. SPC water model [24] was placed in cubic box of 1 Aº; the box was neutralized with Na+ and Cl− ions. Both the systems were heated to 300 K followed by equilibration with NVT for 1 ns at a constant temperature of 300 K. After this step another 1 ns equilibration was performed with NPT-MD. The trajectories generated were subjected to Root mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of gyration (Rg) and Solvent Accessibility Surface Area (SASA) by using inbuilt gromacs tools. g-h bond was used to calculate number of distinct hydrogen bonds formed by specific residues to other amino acids within the protein during the simulation (NH bond).
Principal component analysis
Gromacs 4.5.6 inbuilt tools g-covar and g-anaeig were used for performing Principal Component Analysis (PCA). PCA was used to extract the principal modes involved in the motions of WT and MU prion protein. The Prion protein regions which are accountable for the most significant collective motions can be acknowledged using this analysis.
Visualization
Pymol [25], VMD [26] and Discovery studio visualizer [27] were used for visualization. All the plots were plotted using XMGRACE [28].
Result and Discussion
Structural analysis
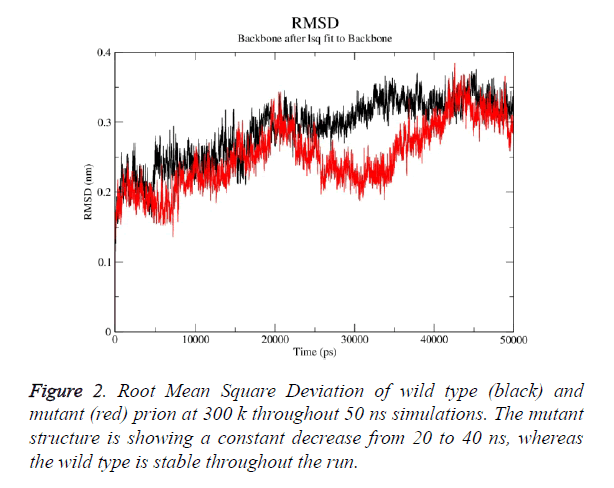
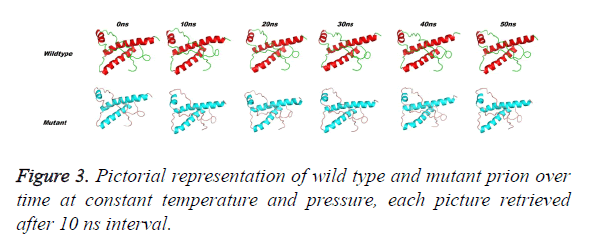
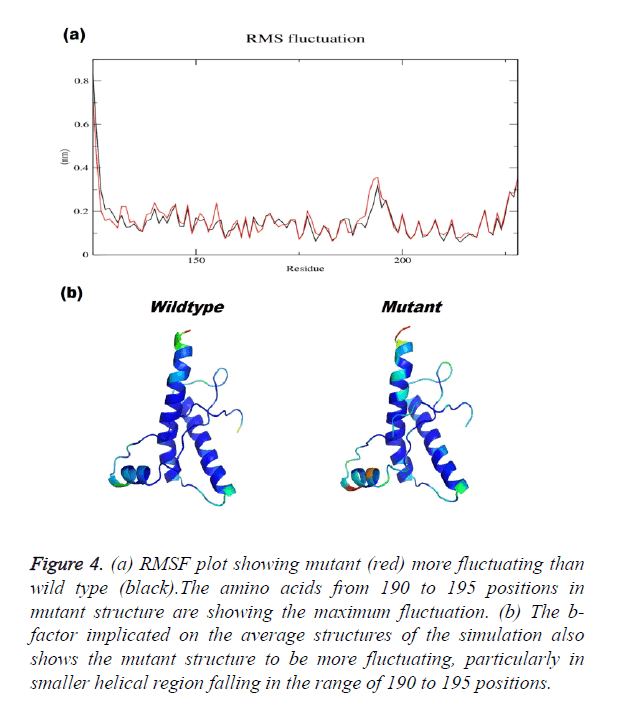
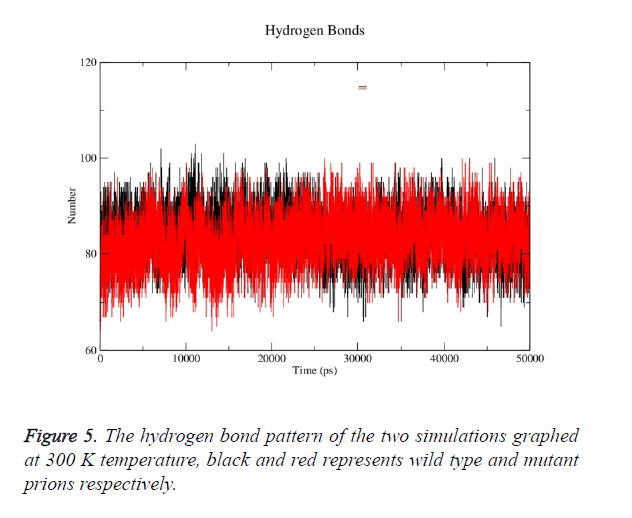
Prion protein ranging from 125-227 amino-acid was subjected to SPDB viewer to look for missing amino acids and energy minimization. The psi-phi distribution shows the acceptability of the overall structure, Figure 1 shows the residues inside and outside the acceptable region and the contour colour represent the hard-sphere and overlap. In both the structures 98% of the residues fall under the permissible region. These structures after 50 ns simulation under GROMOS96 43a1 force field showed stable root mean square deviation at 300 K temperature, both the structure are showing no abnormal variation as shown in Figure 2. Figure 3 shows the picturesque representation of 50 ns run, where each structure is retrieved after 10 ns interval. g-rmsf tool of gromacs was used to compute the root mean square fluctuation (RMSF, i.e. standard deviation) of atomic positions in the trajectories of both wild type and mutant form of prion. The RMSF of the two structures shows the mutant prion structure more fluctuating than wild type as shown in Figure 4a. The fluctuations when subjected onto the average structures of the two simulations and the results are shown in Figure 4b (blue representing the most stable and red most fluctuating). The change in the fluctuation can be because of the decrease in intra-protein hydrogen bond pattern (Figure 5). g-h bond tool was used to compute and analyses hydrogen bonds, hydrogen bonds were determined based on cut-offs for the angle Hydrogen-Donor- Acceptor and the distance Donor-Acceptor. OH and NH groups were regarded as donors, O and N as acceptor by default. Here the calculation was done using a grid of 19x19x19 Aº and the cut off value for hydrogen bond calculation was set at 0.35 Aº. Wild type formed 89 average number of intra-hydrogen bonds per frame, while mutant formed 84 intra-hydrogen bonds per frame. This decrease in number of hydrogen bond in mutant prion is a factor that governs the change in fluctuation, as observed earlier.
Figure 4. (a) RMSF plot showing mutant (red) more fluctuating than wild type (black).The amino acids from 190 to 195 positions in mutant structure are showing the maximum fluctuation. (b) The b-factor implicated on the average structures of the simulation also shows the mutant structure to be more fluctuating, particularly in smaller helical region falling in the range of 190 to 195 positions.
Solvent accessible surface area
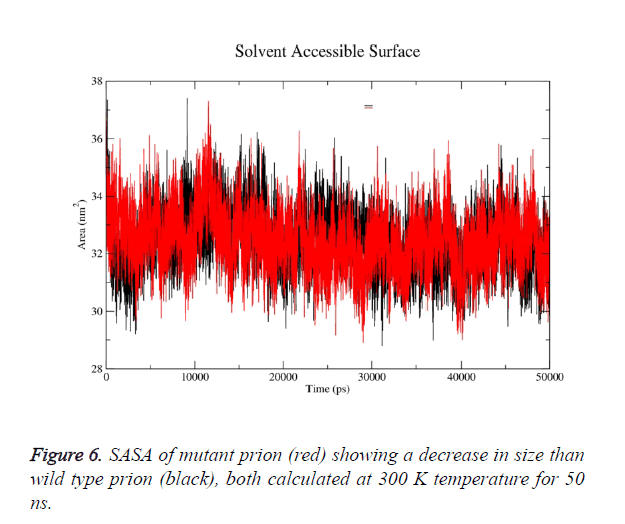
g-sas tool of GROMACS was used to computes hydrophobic, hydrophilic and total Solvent Accessible Surface Area (SASA) of the prion protein variants over time (Figure 6a). The mutant structure has lesser SASA which correlates with its lesser radius of gyration Rg in mutant structure (Figure 6b). g-gyrate tool was used to compute the Rg of a molecule as a function of time. The wild type SASA values fall in the range of 29- 37.5 nm2 and its average over time at 300 K was 33.7 nm2, in mutant the range was 30-36.3 nm2 and its average SASA over run at 32.4 nm2. More than 60% of the difference in SASA is observed in hydrophobic area of prion.
Secondary structure analysis
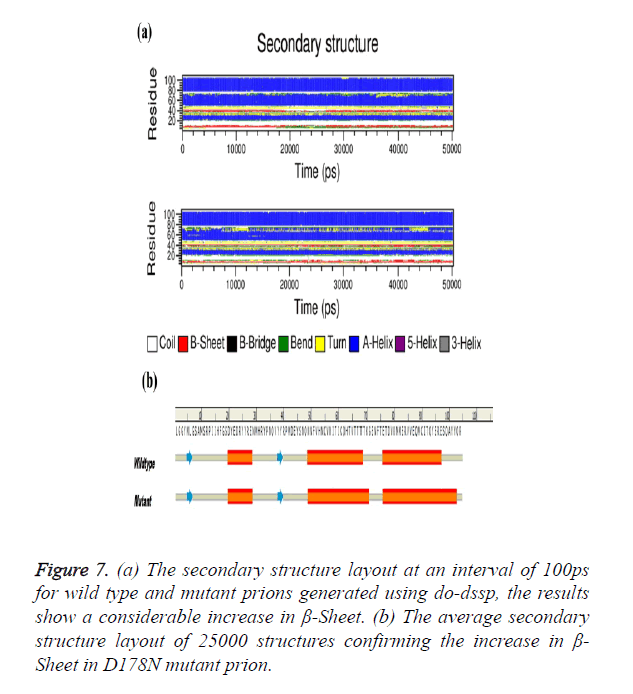
The Secondary structure analysis using do-dssp tool of the two trajectories with each snap shot taken after 100 ps were analysed. Figure 7a shows the change in β-Sheet content of the mutant structure showing considerable presence throughout simulation. The average structure of 25000 structures was selected for secondary structure analysis; mutant average structure of prion is also showing considerable increase in β- sheet (Figure 7b).
Figure 7. (a) The secondary structure layout at an interval of 100ps for wild type and mutant prions generated using do-dssp, the results show a considerable increase in β-Sheet. (b) The average secondary structure layout of 25000 structures confirming the increase in β- Sheet in D178N mutant prion.
Principal component analysis
g-anaeig and g-covar tools were used for PCA. g-anaeig analyses eigenvectors. The eigenvector is derived from covariance matrix. g-covar calculates and diagonalizes the (mass-weighted) covariance matrix. When a trajectory is projected on eigenvectors, all structures are fitted to the structure in the eigenvector file, if present, otherwise to the structure in the structure file. The impact of overall motion of protein was analysed by principal component analysis (PCA) by construction of eigenvectors. For comparing the overall motion in conformational space, PCA plot of the two structures was plotted. In case of complexes, the covariance trace values of wild type and mutant structures were 15 and 13 nm2 respectively. The PCA plot of wild type complex showed the expanded clusters in conformational space. The PCA plot of mutant showed less change in overall motion with clusters decreased in conformational space compared to wild type. Overall, the PCA plot of two structures confirmed the less change in protein motion and clusters decreased compared to wild type complex is due to mutation.
Conclusion
Our study has performed molecular dynamics simulations of a segment (residues 125-228) from HuPrP and its ASP178ASN mutant. The prion protein is prone to structural instability and mutations play an important role in this regard. The mutations lead to instability and conformational plasticity and promote its conversion to intermediate states close to the harmful PrPsc. The structural rearrangement monitored at atomic level as time interval properties has given a detailed insight into the anomalies. These anomalies might act as molecular switch, triggering the conversion of PrPC to PrPsc. The results also show a considerable change in the β-sheet content that can be the leader of the conformational changes and disease-related conversions.
Acknowledgement
The authors would like to thank the management of Hubei xiaogan central hospital infection for their support in the study. Further we like to acknowledge the bioinformatics division of Binzhou Medical University for their support in this study.
References
- Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiological reviews 2008; 88: 673-728.
- Imran M, Mahmood S. An overview of human prion diseases. Virol J 2011; 8: 559-567.
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins Proc Natl Acad Sci U S A 1993; 90: 10962-10966.
- Goldfarb M, Lev G, Brown M, Paul. The transmissible spongiform encephalopathies. Ann rev med 1995; 46: 57-65.
- Mead S, Stumpf MP, Whitfield J, Beck JA, Poulter M, Campbell T, Uphill JB, Goldstein D, Alpers M, Fisher EM, Collinge J. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science 2003; 300: 640-643.
- Gambetti P, Dong Z, Yuan J, Xiao X, Zheng M, Alshekhlee A, Castellani R, Cohen M, Barria MA, Gonzalez-Romero D, Belay ED, Schonberger LB, Marder K, Harris C, Burke JR, Montine T, Wisniewski T, Dickson DW, Soto C, Hulette CM, Mastrianni JA, Kong Q, Zou WQ. A novel human disease with abnormal prion protein sensitive to protease. Annals neurol 2008; 63: 697-708.
- Lugaresi E, Medori R, Montagna P, Baruzzi A, Cortelli P, Lugaresi A, Tinuper P, Zucconi M, Gambetti P. Fatal familial insomnia and dysautonomia with selective degeneration of thalamic nuclei. N Engl J Med 1986; 315: 997-1003.
- Richardson EP Jr, Masters CL. The nosology of Creutzfeldt-Jakob disease and conditions related to the accumulation of PrPCJD in the nervous system. Brain pathol 1995; 5: 33-41.
- Apetri AC, Vanik DL, Surewicz WK. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90-231. Biochemistry 2005; 44: 15880-15888.
- Epstein FH, Haywood AM. Transmissible spongiform encephalopathies. N Engl J Med 1997; 337: 1821-1828.
- Zarranz J, Digon A, Atares B, Rodriguez-Martinez A, Arce A, Carrera N, Fernandez-Manchola I, Fernandez-Martinez M, Fernandez-Maiztegui C, Forcadas I. Phenotypic variability in familial prion diseases due to the D178N mutation. J Neurol Neurosurg Psychiatry 2005; 76: 1491-1496.
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95: 13363-13383.
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216: 136-144.
- Rossetti G, Bongarzone S, Carloni P. Computational studies on the prion protein. Cur topics med chem 2013; 13: 2419-2431.
- Chikan N, Bukhari S, Shabir N, Amin A, Shafi S, Qadri R, Patel T. Atomic Insight into the Altered O6-Methylguanine-DNA Methyltransferase Protein Architecture in Gastric Cancer. PloS one 2014; 10: 0127741
- Zhang WQ, Liu YQ, Ma QS, Wang L, Hu ZH. Effect of Mutation S34N on hCAP in Periodontal Dental Arthritis Patients: Molecular Dynamics Simulation Study. Biomed Res 2015; 26: 771-776.
- Lee S, Antony L, Hartmann R, Knaus KJ, Surewicz K, Surewicz WK, Yee VC. Conformational diversity in prion protein variants influences intermolecular beta-sheet formation. Embo J 2010; 29: 251-262.
- Guex N, Peitsch MC. SwissModel; and the SwissPdb Viewer: an environment for comparative protein modeling. electrophoresis 1997; 18: 2714-2723.
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucl aci res 2003; 31: 3812-3814.
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature methods 2010; 7: 248-249.
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006; 22: 2729-2734.
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009; 25: 2744-2750.
- Hess B, Kutzner C, Van Der Spoel D, Lindahl E. Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of chem th comp. 2008; 4: 435-447.
- Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Physi Chem 2001; 105: 9954-9960.
- DeLano WL. The Pymol molecular graphics system 2002.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J mol graphics 1996; 14: 33-38.
- Studio D. version 3.5. Accelrys Inc: San Diego, CA, USA. 2012.
- Turner P. XMGRACE, Version 5.1.19. Centre for coastal and land-margin research, Oregon Graduate Institute of Science and Technology, Beaverton. Or 2005.