ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2023) Volume 34, Issue 3
Bioinformatics analysis of gene network of thyroid disorders and coronary artery diseases.
Richa Kahol1,2*, Atul Kathait3
1Department of Biosciences, Apeejay Stya University, Gurugram, Haryana, India
2Department of Endocrinology and Diabetes, Medanta the Medicity, Gurugram, Haryana, India
3Amity University, Patna, Bihar, India
- Corresponding Author:
- Richa Kahol
Department of Biosciences
Apeejay Stya University
Gurugram
Haryana
India
Accepted date: June 30, 2023
Background: Thyroid Disorders (TD) and Coronary Artery Diseases (CAD) are among the highly prevalent diseases across the globe. Medical literature suggests a strong interconnection between the occurrences of the two diseases as well as the origin of the two organs. Patients with thyroid abnormalities are likely to develop cardiac dysfunctions and vice versa.
Aim: In this paper we attempt to explore similar biological and genetic features among Thyroid Disorder (TD) and Coronary Artery Diseases (CAD). Secondly, we also analyse the genetic networks to determine the most significant common genes.
Methods: Genes related to the diseases were searched and retrieved from National Centre for Biotechnology Information. Initially 100 genes for thyroid disorders and1896 for coronary artery diseases were downloaded. After pre-processing, we had 94 and 980 exclusively homo-sapiens genes for thyroid disorders and coronary artery diseases, respectively. Also, 37 common genes were explored. Among them 14 significant genes were selected for further analysis. Thereafter, protein-protein interaction network, enrichment analysis, topological properties analysis, physical interaction network, co-expression network, gene regulatory network were constructed. Also, protein-drug interaction, protein-chemical interaction and drug-gene interaction analysis were conducted.
Results: TD and CAD are related to each other on the genetic level. TNF, SIRT1 and STAT3 are the most significant common genes amongst these two diseases.
Conclusion: This analysis establishes a biological and genetic link between the two diseases. Drug-gene analysis indicates some potential drug targets. This can be considered for further verification and validation chemical experiments.
Keywords
Thyroid disorder, Coronary artery diseases, Protein-protein interaction, Gene regulatory networks, Bioinformatics.
Introduction
Cardiovascular Diseases (CVD) are a threat to the mankind globally. Coronary Artery Diseases (CAD) are the most common type of CVD. It is one of the leading causes of death world over as it attributes to 17.8 million deaths annually. In the United States approximately more than 200 million dollars are spent by the healthcare service providers for CAD [1]. About 20.1 million adults of age ≥ 20 have symptomatic CAD (about 7.2% of disease burden). In 2020, about 20% of the deaths caused by CAD were of adults below 65 years of age [2]. CAD has multifactorial aetiology. At least three of the five risk factors of CVD are associated with metabolic syndrome [3]. It is known that thyroid hormone has a wide range of effect on the cardiovascular system. Medically, increase or decrease in the level of thyroid hormone can exacerbate cardiovascular issue ranging from arrhythmias to heart failure or death [4]. Heart functioning is impacted by prevailing comorbidities such as dyslipidemia, arterial hypertension, renal failure and endocrine disorders such as diabetes or thyroid gland disorder. For normal functioning of the heart it is necessary to maintain thyroid hormone level in normal range. TD is commonly observed in subjects with CAD, hypothyroidism being more prevalent as compared to hyperthyroidism [5].
In this study, we explore the relationship between the two diseases on the genetic level. The primary goal is to explore an interactive link among the two dissimilar but clinically relative diseases. In addition, we attempt to form protein-drug/drug-gene interactions using bioinformatics tools. This gives rise to drug design that is applicable in combined treatment of both the diseases. Initially, common genes of the selected diseases were explored and only homo-sapiens genes were selected. These are pre- processed and significant common genes are shortlisted for further analysis. Various bioinformatics tools used throughout the study are mentioned along with their reports. Through this investigation, the significant hub genes for the two diseases are also explored. The explored drug details can be a subject to chemical experimental verification.
Materials and Methods
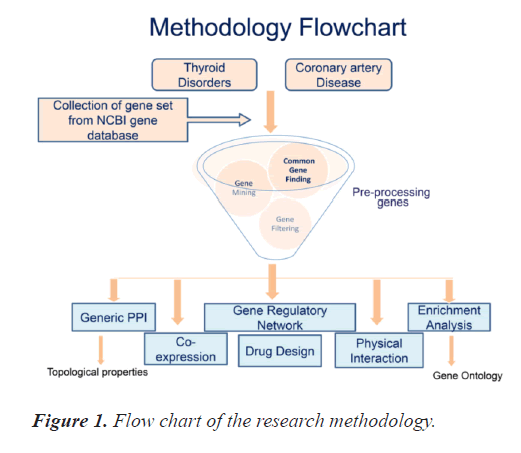
There is paucity of combined bioinformatics based studies for the two diseases included in this research study. This study initiates from collection of the genes of interest and concludes with generation of the PPI and gene regulatory networks for the most significant common genes between TD and CAD. It includes collection and pre-processing of the genes, study of topological properties and gene ontology of the genes as well as drug /chemical interaction using appropriate online bioinformatics tools. Figure 1 exhibits the research methodology's pictorial representation. All the steps included in the study are organised as subsection hereunder.
Gene collection
In this study, gene collection was done using National
Centre for Biotechnology Information (NCBI) officialdatabase. NCBI stores very wide range of various databases including authentic genomic databases. NCBI allows easy download of gene files arranged according to their gene weights. Such gene files were downloaded for all the organisms and specifically for homo-sapiens (as it a human centric study) as well. Thereafter, these files were imported into Microsoft excel for further analysis.
Common genes forage
In this study, identification of candidate genes is an important step, so common genes among the two diseases need to be identified. The Homo sapiens genes for thyroid disorders and coronary artery diseases were processed by data mining. Thereafter, common genes among the two diseases were identified by constructing Venn diagrams. The most significant genes out of these common genes were selected later. The significance of the number of common genes found was statistically justified.
Generic PPI
In this study, after selection of the common genes, their PPI networks were constructed. Generic PPI networks are constructed in order to have a better understanding of the inter-relation between proteins. PPI network enhance the knowledge of genetic signalling pathways of human diseases [6]. In this study we used online bioinformatics tool Network Analyst to construct the generic PPI network. The top most significant genes were selected on the basis of this interactive network.
Topological property
In this study we conducted the Topological Properties (TP) analysis to find the degree of significant gene, clustering coefficient, closeness centrality and between centrality. Initial samples file for topological properties analysis are collected form the PPI networks. For processed TP result we used an online bioinformatics software cytoscape.
Enrichment analysis
In this study we conducted Enrichment Analysis (EA) in order to determine the gene ontology details of the selected gene set. Gene Ontology comprises of molecular functions that gives details of the molecular activities of the genes, Biological processes that gives physiological or cellular details of the genes and cellular components that defines component or organelle of the cell where gene functions (gene products/protiens) are executes. Using the online bioinformatics tool STRING, molecular functions, cellular components and biological process tables were generated.
Co-expression and physical interaction
In this study we generated co-expression network and physical interaction network of the selected significant genes. Co-expression networks are the used to study the process level functionality of the genes at structural level as they are depicted on the basis of pair-wise calculations of gene interaction and their significance levels according to gene expression [7]. On the other hand, physical interaction network explains the gene linkage patterns. Co-expression and physical interaction networks were constructed using online bioinformatics tool Genemania.
Gene Regulatory Network (GRNs)
In this study, we constructed Gene regulatory networks. GRNs provides detailed analysis about the regulatory molecules of a group of genes, hence they are generally build on the basis of different biological functions related to genetic or molecular functionality [8]. Regulators of the gene networks are usually transcription factors, RNAs or metabolites [9]. There different levels of gene regulatory networks, in this study we included TF-gene interaction, Gene-miRNA interaction, and TF-miRNA co-regulatory using Network analyst, an online bioinformatics tool.
Protein drug interaction
In this study, we attempt to design a potential protein- drug target model. With this model we can understand the fundamental characteristics of molecule affinity [10]. The primary requirement of drug design is to decrease its toxicity and to increase its affinity and efficiency [11]. In the present study, the protein-drug target is designed for two diseases using Network analyst, a popular bioinformatics tool.
Protein chemical interaction
In this study, we attempt to design a protein-chemical interaction model. Protein–Chemical interaction calculations are essential for bioinformatics research and are very widely done [7]. Due to the wide range of known datasets and medical literature, specificity of significant chemicals interactions is lacking [12]. Therefore, interaction between chemicals and distinctive target genes are required. For this study, we used Network Analyst generated enrichment analysis.
Drug Gene Interaction (DGI)
In this study, we show the drug gene interactions. Drug- gene interactions are the association between the genetic variant and a potential drug that directly affects the treatment of the patient. For enrichment analysis Network Analyst was used. Using online bioinformatics tools ToppGene and DGIdb, lists of potentially associated drugs and interacting drugs were extracted.
Results
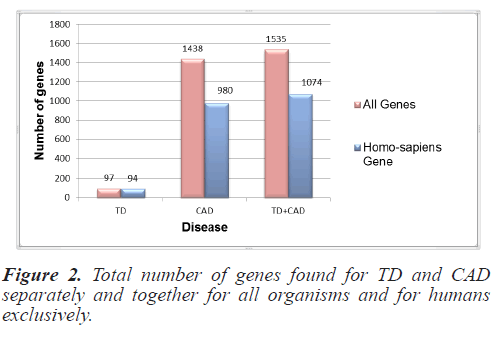
For this study, the genes responsible for TD and CAD were collected from NCBI as it is the most authentic and trusted gene database. Gene sets for all the organisms and specifically for Homo sapiens were collected separately. Initially 100 genes for TD and 1896 for CAD were raised from NCBI files. These get sets were pre-processed and duplicity and redundancy were removed. Finally the gene set for all organisms included 97 genes for TD and 1438 for CAD. Homo sapiens genes included 94 genes and 980 CAD genes. Figure 2 depicts the total number of individual disease genes as well as combined genes.
Common gene finding
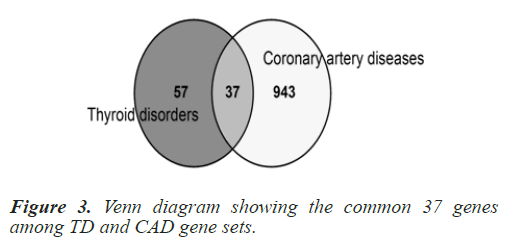
Common genes among selected Homo sapiens gene sets were explored as this a human centric study. To determine the common genes among TD and CAD, a Venn analysis was conducted using online tool ‘Venny’ (https://bioinfogp. cnb.csic.es/tools/venny/). The list of common genes was retrieved from the online tool. 37 common genes were found when the processed Homo sapiens genes of the two disease gene sets (TD+CAD) were overlapped. The common 37 genes are: TNF, IL6, TGFB1, IL10, STAT3, MMP9, HLA-DRB1, IL1B, GSTM1, MMP2, SIRT1, GSTT1, CXCL12, HLA-DQB1, NLRP3, LGALS3, MBL2, CYP1A1, CCL5, TNFRSF1A, NAMPT, FASLG, CALCA, DICER1, IL12B, TIMP2, IL21, UCP2, IL27, TNFSF12, PDE4D, IL5, IL16, COX2, SH2B3, KALRN and ZFAT. In order to cross check the relevance and significance of the explored 37 common genes, a hyper geometric test was run using a hyper geometric p-value calculator. The gene data set was found significant with a p-value of 1.0621e- 27 approx. Table 1 summarizes the hyper geometric test results and Figure 3 shows the number of genes in the venn analysis.
| Hypergeometric test result for 37 common genes | ||
|---|---|---|
| SET 1 | SET 2 | |
| Parameters | 37, 94, 980, 25000 | 37, 980, 94, 25000 |
| Expected number of successes | 3.6848 | 3.6848 |
| Results | over enriched 10.04 fold compared to expectations | over enriched 10.04 fold compared to expectations |
| Hyper geometric p-value | 1.062134375645501e-27 | 1.062134375645501e-27 |
Table 1. Detailed result of statistical hyper geometric test to test significance of 37 common genes among TD and CAD.
Generic PPI
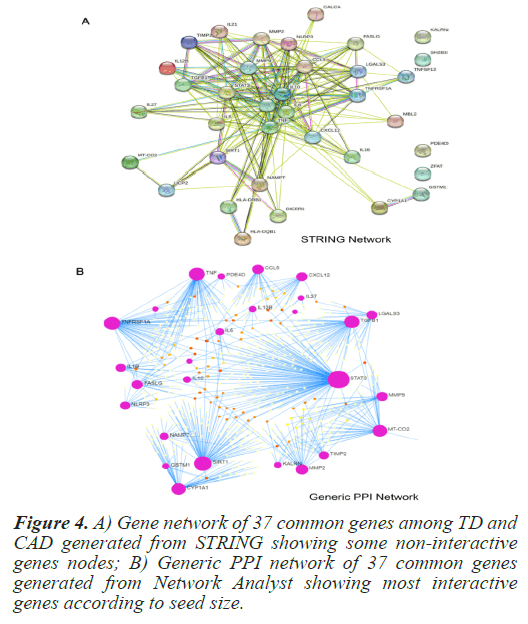
In order to determine direct and indirect link between the selected common genes and hub protiens PPI networks were generated. The STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is a pictorial based biological database that predicts protein-protein interactions. PPI network generated using STRING clearly indicated some non-participating genes. In order to determine most significant interacting genes of the PPI network, Network Analyst was used. Network Analyst delivers data on gene expressions that refer to the Network of Protein-Protein Interactions (PPI) [13]. From the PPI network by Network Analyst, the most significant genes were clearly visible on the basis of their seed size. Figure 4 depicts the PPI network of the 37 common genes from STRING and Network Analyst respectively. From the PPI network, the most significant common genes were selected with degree of interaction higher than or equal to 15. Table 2 shows the degree of top 14 selected genes. In order to check the relevance and significance level of the selected top 14 interacting genes, a hyper geometric test was conducted. These genes were found to significant with the p-value of 1.73e-05. Table 3 summarizes the significance test result.
| Degree of selected 14 genes | ||
|---|---|---|
| Label | Degree | Betweenness |
| STAT3 | 128 | 72311.94 |
| SIRT1 | 67 | 42940.75 |
| TNFRSF1A | 52 | 12790.9 |
| TNF | 47 | 18316.7 |
| TGFB1 | 44 | 19931.75 |
| MT-CO2 | 38 | 16591.38 |
| CYP1A1 | 36 | 18281 |
| MMP2 | 23 | 9952.05 |
| CCL5 | 20 | 7764.03 |
| MMP9 | 18 | 3612.58 |
| CXCL12 | 17 | 5528.36 |
| FASLG | 17 | 4979.43 |
| LGALS3 | 16 | 6583.61 |
| IL1B | 15 | 4737.48 |
Table 2. Degree value of significant top 14 genes (table from network analyst).
| Hypergeometric test result for 14 common genes | ||
|---|---|---|
| SET 1 | SET 2 | |
| Parameters | 37, 94, 980, 25000 | 37, 980, 94, 25000 |
| Expected number of successes | 3.6848 | 3.6848 |
| Results | over enriched 10.04 fold compared to expectations | over enriched 10.04 fold compared to expectations |
| Hypergeometric p-value | 1.062134375645501e-27 | 1.062134375645501e-27 |
Table 3. The significance test of the top 14 common genes with degree ≥ 15 among TD and CAD gene sets of Homo sapiens.
Topological property
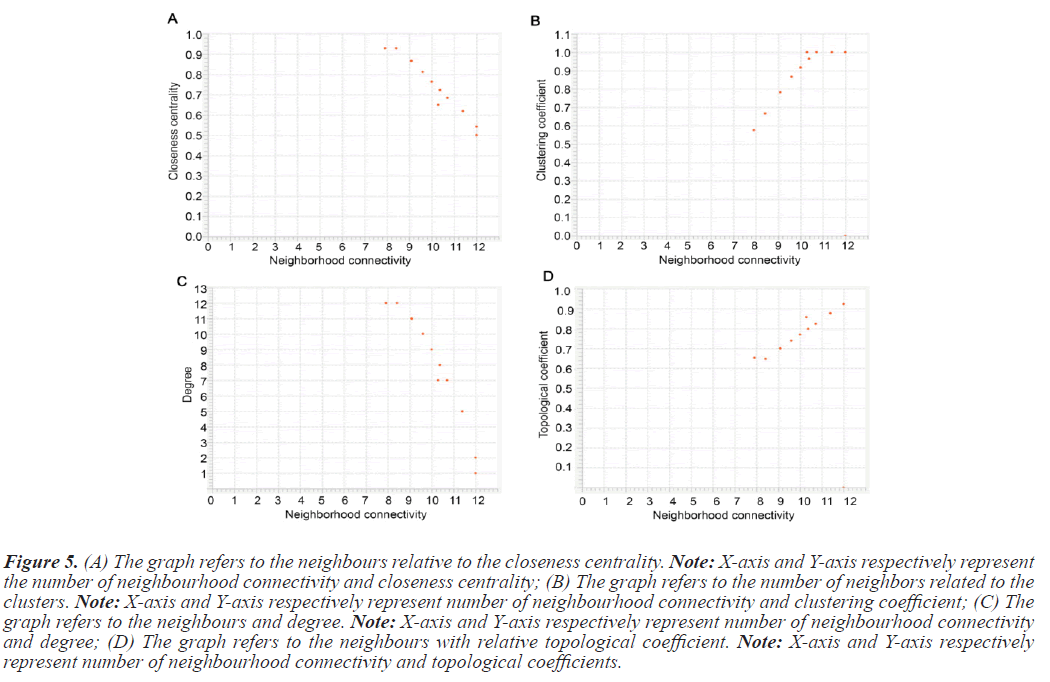
The PPI networks generated in the previous steps were used to determine topological properties. With the help of PPI networks we define specific protein, betweenness centrality, cluster coefficient, topological coefficient, and including degree. The PPI network constructed with STRING was directed send to cytoscape. Table 4 summarizes the value of topological characteristics for 14 seeds. Figure 5 displays the graphical representations of the topological properties.
| Topological properties of 14 genes | ||||||
|---|---|---|---|---|---|---|
| Gene Name | Degree | Neighborhood Connectivity | Clustering Coefficient | Closeness Centrality | Betweenness centrality | Topological Coefficient |
| IL1B | 12 | 0.666666667 | 0.928571429 | 8.416666667 | 0.100702076 | 0.647435897 |
| TNF | 12 | 0.575757576 | 0.928571429 | 7.916666667 | 0.23015873 | 0.652777778 |
| MMP2 | 11 | 0.781818182 | 0.866666667 | 9.090909091 | 0.030189255 | 0.699300699 |
| MMP9 | 11 | 0.781818182 | 0.866666667 | 9.090909091 | 0.030189255 | 0.699300699 |
| STAT3 | 11 | 0.781818182 | 0.866666667 | 9.090909091 | 0.030189255 | 0.699300699 |
| CCL5 | 10 | 0.866666667 | 0.8125 | 9.6 | 0.014163614 | 0.738461538 |
| CXCL12 | 9 | 0.916666667 | 0.764705882 | 10 | 0.007631258 | 0.769230769 |
| TGFB1 | 8 | 0.964285714 | 0.722222222 | 10.375 | 0.001831502 | 0.798076923 |
| TNFRSF1A | 8 | 0.964285714 | 0.722222222 | 10.375 | 0.003663004 | 0.798076923 |
| FASLG | 7 | 1 | 0.65 | 10.28571429 | 0 | 0.857142857 |
| LGALS3 | 7 | 1 | 0.684210526 | 10.71428571 | 0 | 0.824175824 |
| SIRT1 | 5 | 1 | 0.619047619 | 11.4 | 0 | 0.876923077 |
| CYP1A1 | 2 | 1 | 0.541666667 | 12 | 0 | 0.923076923 |
| MT-CO2 | 1 | 0 | 0.5 | 12 | 0 | 0 |
Table 4. Topological properties of 14 seed nodes extracted from cytoscape, network exported from STRING.
Figure 5: (A) The graph refers to the neighbours relative to the closeness centrality. Note: X-axis and Y-axis respectively represent the number of neighbourhood connectivity and closeness centrality; (B) The graph refers to the number of neighbors related to the clusters. Note: X-axis and Y-axis respectively represent number of neighbourhood connectivity and clustering coefficient; (C) The graph refers to the neighbours and degree. Note: X-axis and Y-axis respectively represent number of neighbourhood connectivity and degree; (D) The graph refers to the neighbours with relative topological coefficient. Note: X-axis and Y-axis respectively represent number of neighbourhood connectivity and topological coefficients.
Enrichment analysis
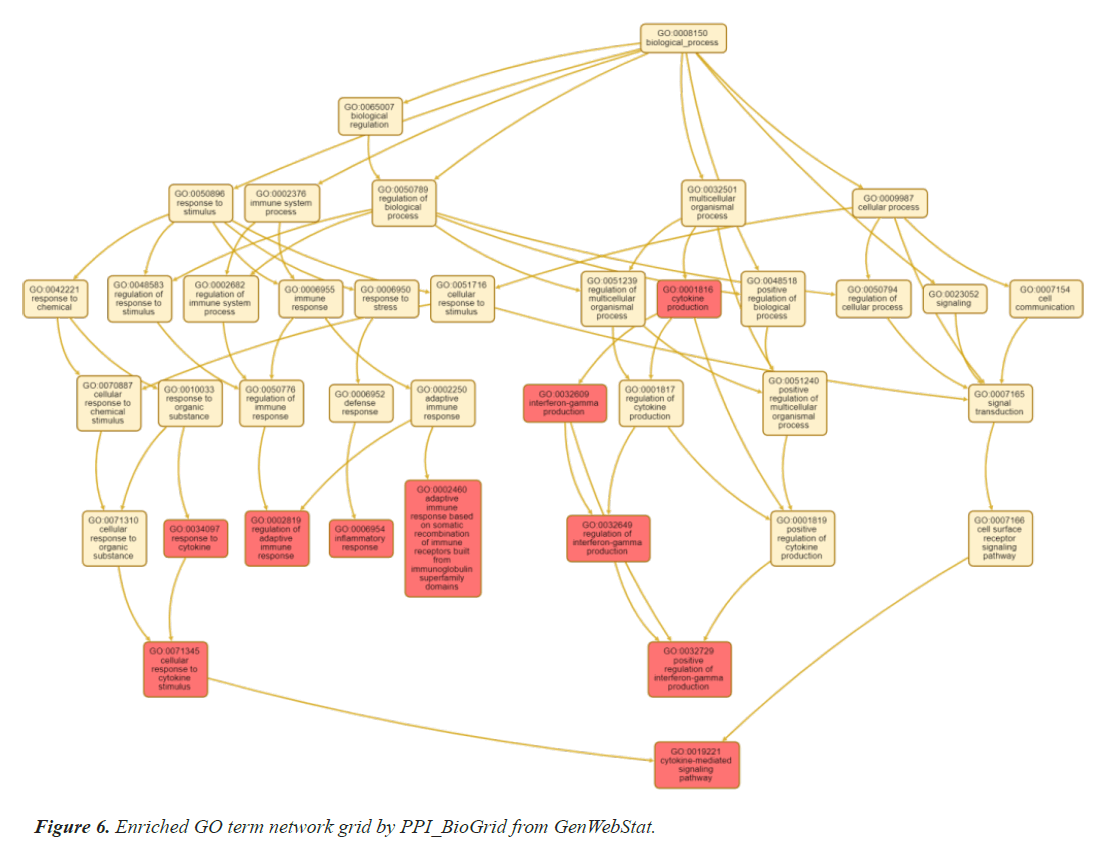
The selected genes are further analysed by gene enrichment. For this gene ontology is studied in detail, which comprise of molecular function, biological process, and cellular component. Gene ontology is one of the most essential features of enrichment analysis which is drafted by STRING, an online bioinformatics tool. Tables 5 and 6 summarizes the result of gene ontology studies, human phenotype and disease-gene association analysis respectively using STRING. These clearly indicate the associations with thyroid gland and heart related diseases. Online bioinformatics software WebGenStat was also employed to fetch an enriched GO term grid meshwork. Figure 6 represents the grid formed by the significant 14 genes.
| Enrichment analysis: Gene ontology for 14 significant common genes | |||||
|---|---|---|---|---|---|
| Biological Process (GO) | |||||
| GO term | Description | Observed gene count | False discovery | ||
| GO:0019221 | Cytokine-mediated signalling pathway | 11 | 678 | ||
| GO:0030335 | Positive regulation of cell migration | 9 | 522 | ||
| GO:0006950 | Response to stress | 14 | 3485 | ||
| GO:0008284 | Positive regulation of cell population proliferation | 10 | 919 | ||
| GO:2001234 | Negative regulation of apoptotic signalling pathway | 7 | 232 | ||
| GO:0070887 | Cellular response to chemical stimulus | 13 | 2919 | ||
| GO:1901700 | Response to oxygen-containing compound | 11 | 1567 | ||
| GO:0042127 | Regulation of cell population proliferation | 11 | 1642 | ||
| GO:0010941 | Regulation of cell death | 11 | 1696 | ||
| GO:0071310 | Cellular response to organic substance | 12 | 2369 | ||
| GO:0044419 | Interspecies interaction between organisms | 11 | 1899 | ||
| GO:0060548 | Negative regulation of cell death | 9 | 999 | ||
| GO:0071677 | Positive regulation of mononuclear cell migration | 4 | 27 | ||
| GO:0097191 | Extrinsic apoptotic signalling pathway | 5 | 100 | ||
| GO:0019221 | Cytokine-mediated signalling pathway | 11 | 678 | ||
| Molecular Functions (GO) | |||||
| GO term | Description | Observed gene count | False discovery | ||
| GO:0005126 | Cytokine receptor binding | 7 | 9.21E-07 | ||
| GO:0005125 | Cytokine activity | 6 | 1.39E-05 | ||
| GO:0048018 | Receptor ligand activity | 7 | 2.07E-05 | ||
| GO:0005102 | Signalling receptor binding | 9 | 0.00011 | ||
| GO:0031730 | CCR5 chemokine receptor binding | 2 | 0.0079 | ||
| GO:0042379 | Chemokine receptor binding | 3 | 0.0079 | ||
| GO:0098772 | Molecular function regulator | 9 | 0.0474 | ||
| GO:0042802 | Identical protein binding | 7 | 0.0486 | ||
| Cellular Component (GO) | |||||
| GO term | Description | Observed gene count | False discovery | ||
| GO:0005615 | Extracellular space | 10 | 0.0103 | ||
| GO:0009986 | Cell surface | 6 | 0.0103 | ||
| GO:0019866 | Organelle inner membrane | 5 | 0.0103 | ||
| GO:0062023 | Collagen-containing extracellular matrix | 5 | 0.0103 | ||
| GO:0005739 | Mitochondrion | 7 | 0.015 | ||
Table 5. Gene enrichment analysis results of 14 significant genes for biological processes, molecular functions and cellular components using STRING.
| Human Phenotype (Monarch) String | ||||
|---|---|---|---|---|
| GO term | Description | Observed gene count | False discovery | |
| HP:0003040 | Arthropathy | 3 | 0.0094 | |
| HP:0100651 | Type I diabetes mellitus | 3 | 0.0094 | |
| HP:0002315 | Headache | 4 | 0.0378 | |
| HP:0010783 | Erythema | 3 | 0.0383 | |
| HP:0012649 | Increased inflammatory response | 6 | 0.0383 | |
| HP:0000820 | Abnormality of the thyroid gland | 4 | 0.0426 | |
| HP:0002715 | Abnormality of the immune system | 7 | 0.0426 | |
| HP:0002960 | Autoimmunity | 3 | 0.0426 | |
| HP:0006530 | Abnormal pulmonary interstitial morphology | 3 | 0.0467 | |
| HP:0001744 | Splenomegaly | 4 | 0.049 | |
| Disease-gene association (disease) STRING | ||||
| GO term | Description | Observed gene count | False discovery | |
| DOID:0050828 | Artery disease | 4 | 0.0029 | |
| DOID:2914 | Immune system disease | 6 | 0.0029 | |
| DOID:438 | Autoimmune disease of the nervous system | 3 | 0.0034 | |
| DOID:11396 | Pulmonary edema | 2 | 0.0044 | |
| DOID:612 | Primary immunodeficiency disease | 5 | 0.0044 | |
| DOID:7 | Disease of anatomical entity | 11 | 0.0052 | |
| DOID:417 | Autoimmune disease | 4 | 0.0107 | |
| DOID:1936 | Atherosclerosis | 2 | 0.0141 | |
| DOID:326 | Ischemia | 2 | 0.0193 | |
| DOID:2377 | Multiple sclerosis | 2 | 0.0202 | |
Table 6. TD and CAD related human phenotype and disease-gene association analysis using STRING.
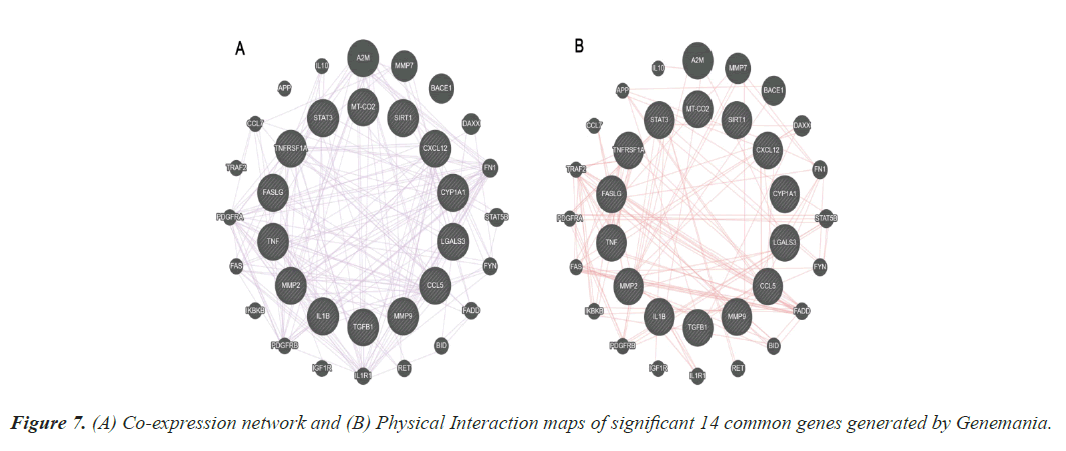
Co expression and physical interaction
The results from the topological properties analysis and the enrichment analysis of the 14 significant genes were further strengthened by the co-expression and physical interaction maps. These maps were generated using Genemania, an online bioinformatics tools. Figure 7 represents the co-expression and physical maps of the 14 significant common genes among TD and CAD.
Gene regulatory network
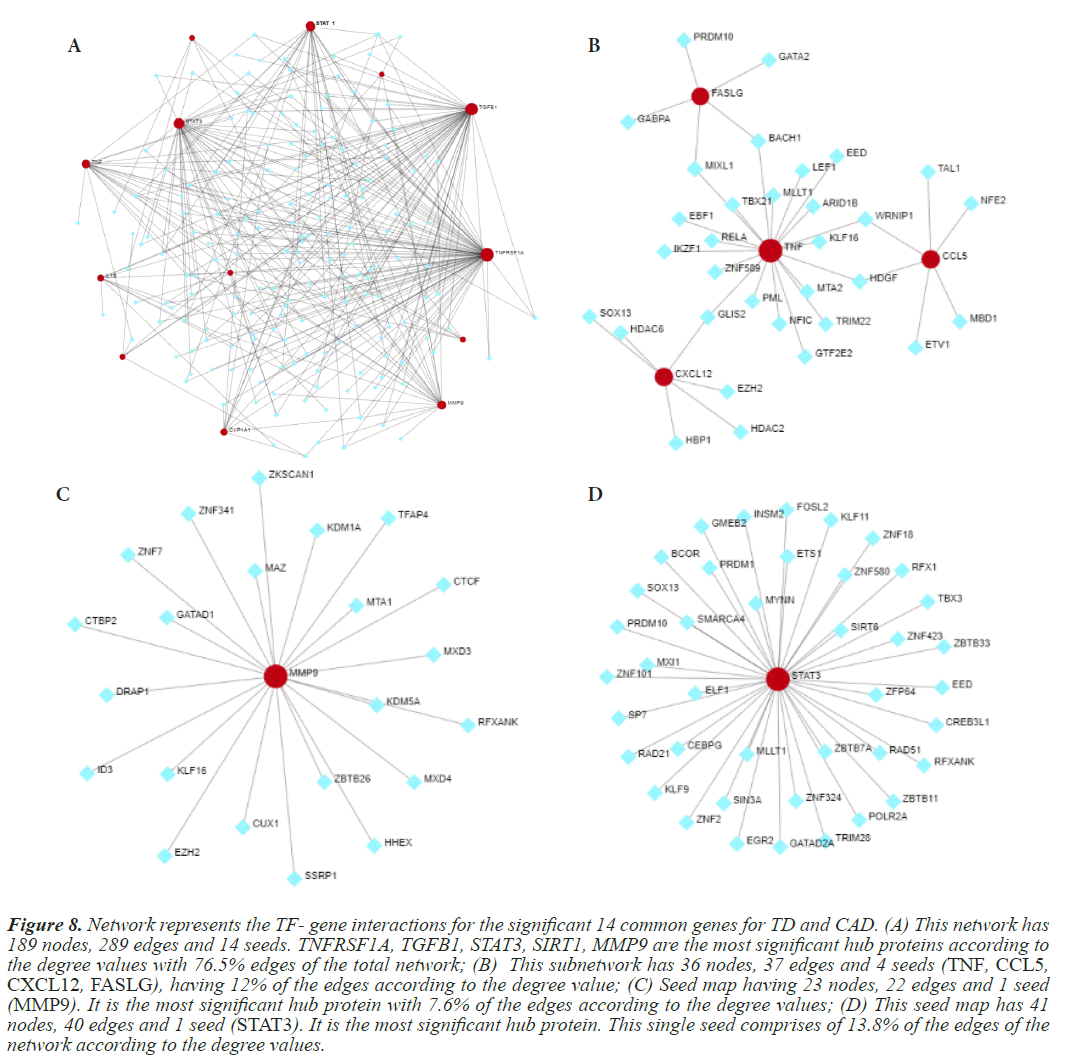
Further confirmation of the TD and CAD interacting genes is done by the gene regulatory networks. In this analysis, three GRNs are designed a TF-gene interaction network, Gene-miRNA interaction and TF-miRNA co-regulatory network. These GRNs are constructed using online bioinformatics tool, Network Analyst, wherein the genes of interest, termed as “seeds” from the previous analysis are mapped to the corresponding molecular interaction databases. Analysis will result in a big sub network, termed as “continent” and some smaller sub networks, termed as “island”. For TF-Gene interaction network, transcription factors and gene target data are derived from the ENCODE ChIP-seq data. For miRNA-gene interaction network data is collected from miRTarBase, which is comprehensive and experimentally validated. For the TF-miRNA co-regulatory network generation RegNetwork repository is employed to collect literature curated regulatory interaction information. It is applicable to human and mouse data exclusively. Figures 8-11 depict the gene regulatory networks.
Figure 8: Network represents the TF- gene interactions for the significant 14 common genes for TD and CAD. (A) This network has 189 nodes, 289 edges and 14 seeds. TNFRSF1A, TGFB1, STAT3, SIRT1, MMP9 are the most significant hub proteins according to the degree values with 76.5% edges of the total network; (B) This subnetwork has 36 nodes, 37 edges and 4 seeds (TNF, CCL5, CXCL12, FASLG), having 12% of the edges according to the degree value; (C) Seed map having 23 nodes, 22 edges and 1 seed (MMP9). It is the most significant hub protein with 7.6% of the edges according to the degree values; (D) This seed map has 41 nodes, 40 edges and 1 seed (STAT3). It is the most significant hub protein. This single seed comprises of 13.8% of the edges of the network according to the degree values.
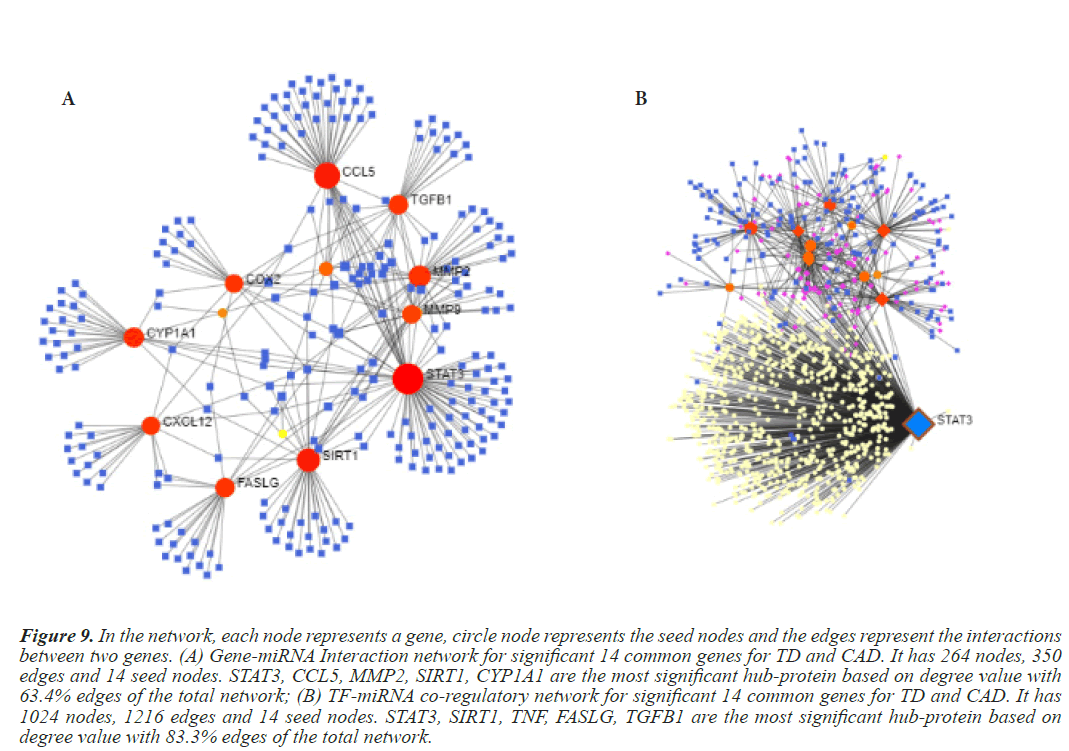
Figure 9: In the network, each node represents a gene, circle node represents the seed nodes and the edges represent the interactions between two genes. (A) Gene-miRNA Interaction network for significant 14 common genes for TD and CAD. It has 264 nodes, 350 edges and 14 seed nodes. STAT3, CCL5, MMP2, SIRT1, CYP1A1 are the most significant hub-protein based on degree value with 63.4% edges of the total network; (B) TF-miRNA co-regulatory network for significant 14 common genes for TD and CAD. It has 1024 nodes, 1216 edges and 14 seed nodes. STAT3, SIRT1, TNF, FASLG, TGFB1 are the most significant hub-protein based on degree value with 83.3% edges of the total network.
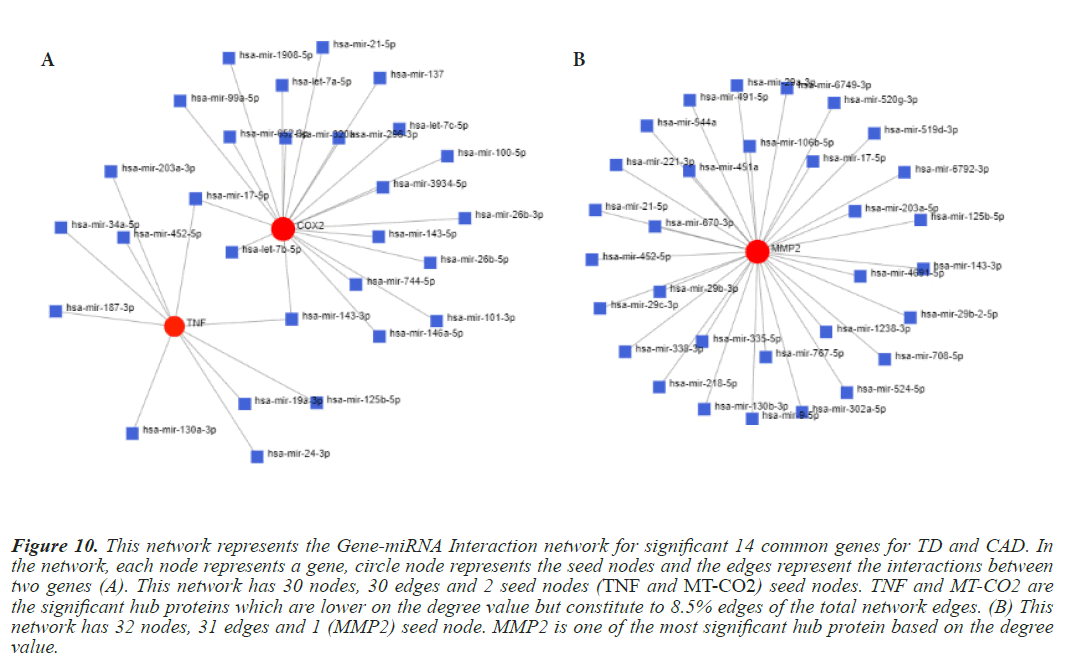
Figure 10: This network represents the Gene-miRNA Interaction network for significant 14 common genes for TD and CAD. In the network, each node represents a gene, circle node represents the seed nodes and the edges represent the interactions between two genes (A). This network has 30 nodes, 30 edges and 2 seed nodes (TNF and MT-CO2) seed nodes. TNF and MT-CO2 are the significant hub proteins which are lower on the degree value but constitute to 8.5% edges of the total network edges. (B) This network has 32 nodes, 31 edges and 1 (MMP2) seed node. MMP2 is one of the most significant hub protein based on the degree value.
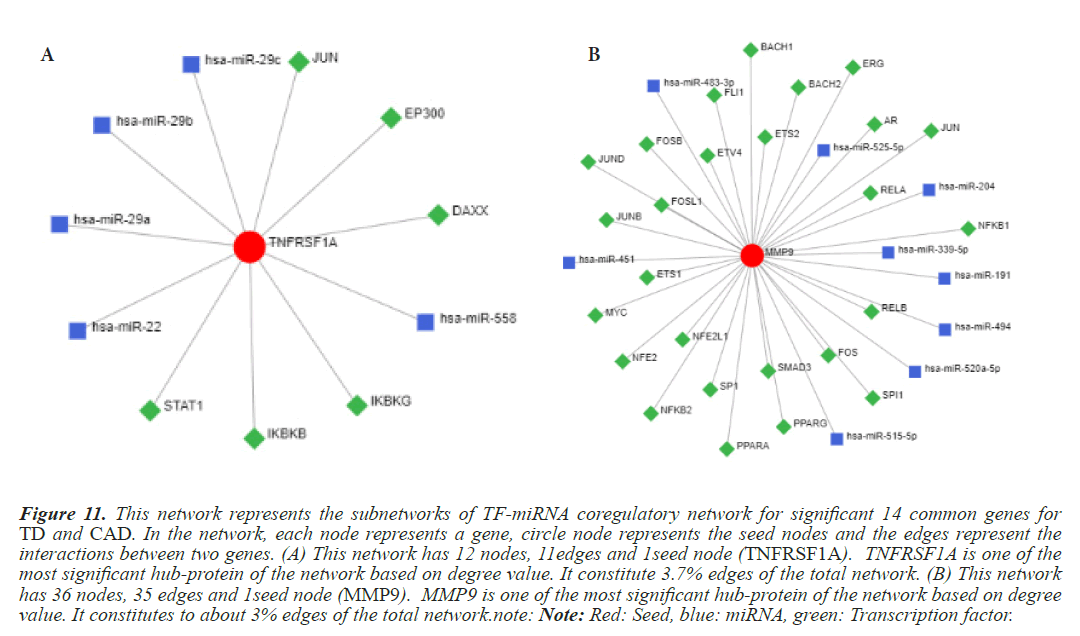
Figure 11: This network represents the subnetworks of TF-miRNA coregulatory network for significant 14 common genes for TD and CAD. In the network, each node represents a gene, circle node represents the seed nodes and the edges represent the interactions between two genes. (A) This network has 12 nodes, 11edges and 1seed node (TNFRSF1A). TNFRSF1A is one of the most significant hub-protein of the network based on degree value. It constitute 3.7% edges of the total network. (B) This network has 36 nodes, 35 edges and 1seed node (MMP9). MMP9 is one of the most significant hub-protein of the network based on degree value. It constitutes to about 3% edges of the total network.note: Note: Red: Seed, blue: miRNA, green: Transcription factor.
Protein Drug Interaction (PDI)
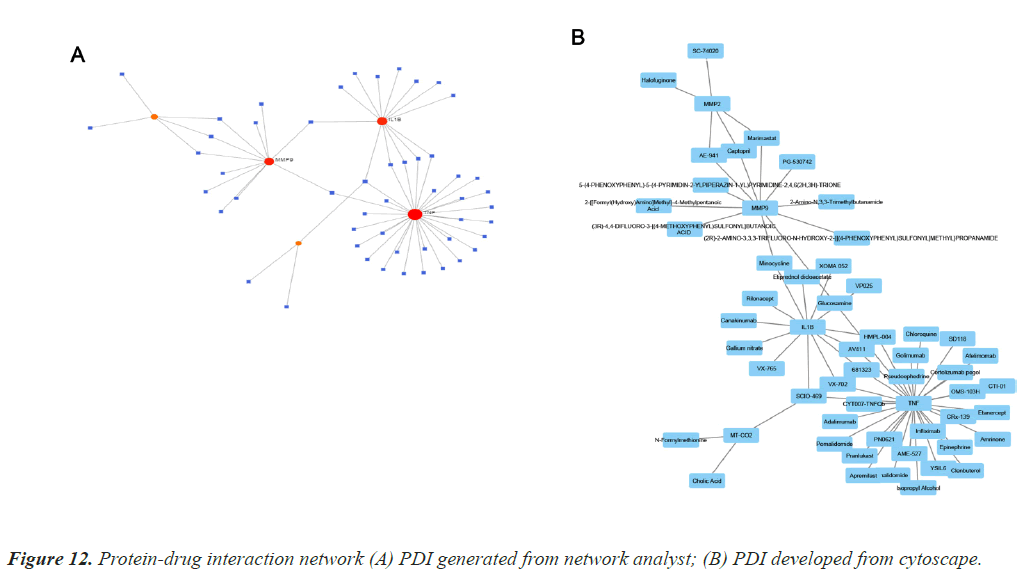
Protein-drug interaction is highly important to clearly understand the basic feature of molecule affinity [10]. Computational methods are generally employed to determine the protein targets for drugs and vice-versa [6]. The interaction information used to develop the PDI map is collected from Drug Bank databases, it is exclusively applicable to human data. Figure 12 depicts the two PDI networks. Interaction network for TD and CAD is generated and developed using Network Analyst and Cytoscape.
Protein Chemical Interaction (PCI)
In this study Protein-chemical interactions are done to observe the versatility of the seed of interest. PCI is an integral part of bioinformatics research as these interactions impact the biological processes of living beings [10]. The interaction information used to develop the PDI map is collected from Comparitive Taxonomics Databases (CTD). PCI maps were generated using NetworkAnalyst. Inidvidual seed PCI maps are compiled in Figure 13.
Figure 13: Protein-chemical interaction network. Each seed has nodes and edges, these are explained here. TNF (node: 956, seed: 955), ILIB (node: 617, seed: 616), STAT3 (node: 219, seed: 218), CCL5 (node: 193, seed: 192), CXCL12 (node: 104, seed: 103), TNFRSF1A (node: 98, seed: 97), MT-COX2 (node: 26, seed: 25), CYP1A1 (node: 689, seed: 688), MMP9 (node: 374, seed: 373), MMP2 (node: 280, seed: 279), TGFB1 (node: 239, seed: 238), FASLG (node: 168, seed: 167), SIRT3 (node: 219, seed: 218), LGALS3 (node: 58, seed: 57).
Drug Gene Interaction (DGI)
In this study, drug gene interactions were explored. For ToppGene online tool, the significant 14 common genes were entered and run. This tool returned the analysed list of the possibly potential drug and its details. Table 7 compiles the details of the relevant parts of the entire result extracted. Similarly, DGIdb also returned a list of potential drug along with the target gene name. Table 8 tabulate the top 5 precise potential drugs extracted using updated online database repositories. This gives a basic interaction map for the development of one drug to target the involved diseases and also work at the molecular level with higher specificity. Tables 7 and 8 summarize the details.
| ToppGene analysis of drug-gene network | ||||||
|---|---|---|---|---|---|---|
| S.no | ID | Name | Source | p-value | Gene from input | Gene in annotation |
| 1 | ctd:D017382 | Reactive oxygen species | CTD | 5.19E-21 | 12 | 325 |
| 2 | ctd:D012431 | Rutin | CTD | 1.31E-18 | 9 | 105 |
| 3 | ctd:D005978 | Glutathione | CTD | 2.37E-18 | 11 | 339 |
| 4 | ctd:D015232 | Dinoprostone | CTD | 7.03E-18 | 10 | 225 |
| 5 | ctd:C093642 | SB 203580 | CTD | 1.06E-17 | 11 | 388 |
| 6 | ctd:D008550 | Melatonin | CTD | 1.54E-17 | 10 | 243 |
| 7 | ctd:C090942 | 4-(4-fluorophenyl)-2-(4-hydroxyphenyl)-5-(4-pyridyl)imidazole | CTD | 1.55E-17 | 9 | 137 |
| 8 | ctd:C017803 | zinc protoporphyrin | CTD | 2.61E-17 | 9 | 145 |
| 9 | ctd:C005274 | naringin | CTD | 2.78E-17 | 9 | 146 |
| 10 | ctd:C007095 | cobaltiprotoporphyrin | CTD | 6.11E-17 | 9 | 159 |
| 11 | ctd:C070081 | fulvestrant | CTD | 1.23E-16 | 11 | 484 |
| 12 | ctd:C432165 | pyrazolanthrone | CTD | 2.65E-16 | 10 | 322 |
| 13 | ctd:D009569 | Nitric Oxide | CTD | 4.08E-16 | 10 | 336 |
| 14 | ctd:C031927 | hydroquinone | CTD | 5.25E-16 | 9 | 201 |
| 15 | ctd:D015735 | Mifepristone | CTD | 5.37E-16 | 11 | 553 |
| 16 | ctd:C434003 | 3-(4-methylphenylsulfonyl)-2-propenenitrile | CTD | 8.39E-16 | 8 | 113 |
| 17 | ctd:C007517 | diphenyleneiodonium | CTD | 9.02E-16 | 8 | 114 |
| 18 | CID000005056 | trans-3,4,5-trihydroxystilbene | Stitch | 1.53E-15 | 11 | 608 |
| 19 | ctd:C107773 | pterostilbene | CTD | 2.65E-15 | 8 | 130 |
| 20 | ctd:D002101 | Cacodylic Acid | CTD | 5.67E-15 | 9 | 261 |
| 21 | ctd:C433788 | lipopolysaccharide, E. coli O26-B6 | CTD | 6.12E-15 | 8 | 144 |
| 22 | CID000145068 | nitric oxide | Stitch | 9.65E-15 | 12 | 1075 |
| 23 | ctd:D003035 | Cobalt | CTD | 9.74E-15 | 9 | 277 |
| 24 | ctd:D013311 | Streptozocin | CTD | 1.70E-14 | 11 | 757 |
| 25 | ctd:D019808 | Losartan | CTD | 1.86E-14 | 8 | 165 |
Table 7. Top 25 outcomes of drug-gene interactions map results from Toppgene analysis.
| DGIdb high affinity drugs for gene of interest | ||
|---|---|---|
| TNF (definite matches) | ||
| Drug | Interaction | Source |
| ONERCEPT | Inhibitor | ChemblInteractions |
| AZ9773 | Inhibitor | ChemblInteractions |
| VADIMEZAN | Inducer | TALC |
| TNFRSF1A(definite matches) | ||
| Drug | Interaction | Source |
| INFLIXIMAB | PharmGKB | |
| ETANERCEPT | PharmGKB | |
| CYCLOPHOSPHAMIDE | NCI | |
| STAT3 (definite matches) | ||
| Drug | Interaction | Source |
| AZD-1480 | DTC | |
| ACITRETIN | TTD | |
| CIGLITAZONE | DTC | |
| SIRT1 (definite matches) | ||
| Drug | Interaction | Source |
| CAMBINOL | Inhibitor | TTD |
| CHEMBL375563 | DTC | |
| CHEMBL200762 | DTC | |
| TGFB1 (definite matches) | ||
| Drug | Interaction | Source |
| FRESOLIMUMAB | Inhibitor/antibody | TALC|ChemblInteractions |
| METELIMUMAB | Inhibitor | ChemblInteractions |
| LY-2382770 | Inhibitor | ChemblInteractions |
| MT-CO2 (definite matches) | ||
| Drug | Interaction | Source |
| CELECOXIB | DTC | |
| DACARBAZINE | PharmGKB | |
| GRANISETRON | PharmGKB | |
| MMP2 (definite matches) | ||
| Drug | Interaction | Source |
| MARIMASTAT | Inhibitor | TdgClinicalTrial|TEND|TTD |
| S-3304 | Vaccine | TALC |
| PRINOMASTAT | Vaccine | TALC|TTD |
| MMP9 (definite matches) | ||
| Drug | Interaction | Source |
| MARIMASTAT | Inhibitor | TdgClinicalTrial|TEND |
| PRINOMASTAT | Vaccine | TALC |
| ANDECALIXIMAB | Inhibitor|antibody | ChemblInteractions|TTD |
| CXCL12 (definite matches) | ||
| Drug | Interaction | Source |
| VINCRISTINE | PharmGKB | |
| ALEMTUZUMAB | PharmGKB | |
| CHLORAMBUCIL | PharmGKB | |
| FASLG (definite matches) | ||
| Drug | Interaction | Source |
| ASUNERCEPT | TdgClinicalTrial | |
| LGALS3 (definite matches) | ||
| Drug | Interaction | Source |
| LACTOSE, ANHYDROUS | DTC | |
| BELAPECTIN | TTD | |
| IL1B (definite matches) | ||
| Drug | Interaction | Source |
| INFLIXIMAB | PharmGKB | |
| ASPIRIN | PharmGKB | |
| RILONACEPT | Inhibitor|binder | ChemblInteractions|TTD |
Table 8. Top three high affinity drugs for the target genes, results extracted from DGIdb analysis.
Discussion
The aim of this research study was to explore a genetic link among the selected diseases that are TD and CAD. To achieve this goal, genes of the two diseases were searched, retrieved and collected from NCBI. A large number of gene data sets were retrieved. Thereafter only Homo sapiens genes were selected, which were further pre-processed, filtered and mined. Common genes of the two diseases were found. Common genes were found to be 37 (TNF, IL6, TGFB1, IL10, STAT3, MMP9, HLA-DRB1, IL1B, GSTM1, MMP2, SIRT1, GSTT1, CXCL12, HLA-DQB1, NLRP3, LGALS3, MBL2, CYP1A1, CCL5, TNFRSF1A, NAMPT, FASLG, CALCA, DICER1, IL12B, TIMP2, IL21, UCP2, IL27, TNFSF12, PDE4D, IL5, IL16, COX2, SH2B3, KALRNand ZFAT). Out of these 37 common genes most significant 14 genes (TNF, TGFB1, STAT3, MMP9, IL1B, MMP2, SIRT1, CXCL12, LGALS3, CYP1A1, CCL5, TNFRSF1A, FASLG and COX2) were selected on the basis of the degree value and the statistical level of significance. The genetic analysis of these significant 14 common genes was done. To ensure interaction among the significant 14 common genes, generic PPI network was constructed using Network Analyst and analysed using Cytoscape. The topological properties of the network were analysed with help of correlations between clustering coefficient, closeness centrality, degree, topological coefficient and neighbourhood connectivity. With the gene ontology of the enriched genes, molecular functions, biological functions and cellular components details of the significant genes were retrieves. In order to ensure the association of the genes at system level functionality, co-expression and physical interaction network were generated with the help of genemania to determine the functional analysis of genomic patterns, gene regulatory networks for TF-gene, gene-miRNA, miRNA-TF interactions were generated using Network Analyst. To check the interaction of the target genes with various proteins and drugs, protein- drug and protein-chemical interaction network maps were created. To understand the association of the target genes with the drugs, drug-gene interactions were analysed in detail. The detailed analysis was done with the help of online tools, ToppGene and DGIdb.
Conclusion
In this research we correlated two dissimilar but clinically relative diseases (TD and CAD) at the gene level. The common genes among these two diseases were explored and pre-processed. Only Homo sapiens specific genes were selected. From the processed 37 common genes, significant 14 common genes were selected (on the basis of the degree value) for further analysis. With these genes, gene topology and gene ontology were studied, generic PPI networks, gene regulatory networks, co-expression and physical interaction networks, protein-chemical, protein- drug and drug gene association networks were constructed. With the help of all these analysis and detailed studies, finally we explored the most interactive and significant 5 genes (TNF, SIRT1 and STAT3). These are also termed as the hub proteins. Although the primary goal of this study was to explore and correlate the common genes among the two diseases but this bioinformatics based study forms a platform for gene targeted drug design.Declarations
Ethical approval
Not applicable
Competing interests
Authors declare no competing interests of any nature.
Author’s contribution
Richa Kahol has conceived and designed the study, performed research and analysed the data. Also wrote and reviewed the paper. Atul Kathait has conceived and designed the study. Also, reviewed the paper
Funding
No funding is received for this study.
Availability of data and materials
The methodology clearly explains the websites and online tools applicable.
References
- Brown JC, Gerhardt TE, Kwon E. Risk factors for coronary artery disease. StatPearls 2022.
[Google Scholar] [PubMed]
- Tsao CW, Aday AW, Almarzooq ZI. Heart disease and stroke statistics-2022 update: A report from the American heart association. Circulation 2022;145:e153-e639.
[Google Scholar] [PubMed] [Crossref]
- Rarau P, Pulford J, Gouda H. Socio-economic status and behavioural and cardiovascular risk factors in Papua New Guinea: A cross-sectional survey. PLoS ONE 2019;14:e0211068.
[Google Scholar] [PubMed] [Crossref]
- Cappola AR, Desai AS, Medici M. Thyroid and cardiovascular disease: Research agenda for enhancing knowledge, prevention, and treatment. Thyroid 2019;29:760-777.
[Google Scholar] [PubMed] [Crossref]
- Richa K, Atul K, Sunil KM. How widespread is thyroid disorders among patients undergoing coronary artery bypass grafting? A study from tertiary care hospital. Asian Pac J Health Sci 2022;9:27-31.
[Crossref]
- Hasan MR, Paul BK, Ahmed K. Computational analysis of network model based relationship of mental disorder with depression. Biointerface Res Appl Chem 2020;10:6293-6305.
- Kawsar Md, Tasnimul AT, Bikash KP. Analysis of gene network model of thyroid disorder and associated diseases: A bioinformatics approach. Informatics in Medicine Unlocked 2020;20:100381.
- Yu D, Lim, J, Wang X. Enhanced construction of gene regulatory networks using hub gene information. BMC Bioinformatics 2017;18:1-20.
[Google Scholar] [PubMed] [Crossref]
- Taz TA, Kawsar M, Paul BK. Computational analysis of regulatory genes network pathways among devastating cancer diseases. J Proteomics 2020;11:63-76.
- Hasan MR, Paul BK, Ahmed K. Design protein-protein interaction network and protein-drug interaction network for common cancer diseases: A bioinformatics approach. Informatics in Medicine Unlocked 2020;18:100311.
- Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput Biol 2012;8:e1002375.
[Google Scholar] [PubMed] [Crossref]
- Kuhn M, Szklarczyk D, Franceschini A. STITCH 3: Zooming in on protein-chemical interactions. Nucleic Acids Res 2012;40:D876-D880.
[Google Scholar] [PubMed] [Crossref]
- Zhou G, Soufan O, Ewald J. Network analyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47:W234-W241.
[Google Scholar] [PubMed] [Crossref]