ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Case Report - Biomedical Research (2018) Volume 29, Issue 10
Clinical characteristics and gene mutation analysis of one pedigree with glycogen storage disease type IV
1Department of Pediatrics, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, PR China
2Department of Pediatrics, the First Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, PR China
- *Corresponding Author:
- Sinian Pan
Department of Pediatrics
The Third Affiliated Hospital of Sun Yat-sen University, PR China
Accepted on April 9, 2018
DOI: 10.4066/biomedicalresearch.29-18-267
Visit for more related articles at Biomedical ResearchGlycogen Storage Disease type IV (GSD-IV; also known as Andersen disease) is a rare autosomal recessive disorder of glycogen metabolism caused by mutations in the Glycogen-Branching Enzyme 1 (GBE1) gene that encodes the 1,4-α-glucan-branching enzyme. The aim of this study is to identify the mutation of GBE1 gene in a family with GSD IV. A 17-month-old Chinese girl with hepatosplenomegaly was admitted to the Third Affiliated Hospital of Sun Yat-sen University (Guangzhou, China). She had shown signs of jaundice and failure to thrive. The peripheral blood samples from the proband and her parents and two older brothers were collected, and the pathogenic genes and mutational sites of the proband were screened by the next generation sequencing and verified by Sanger sequencing. In the family, the proband carried a homozygous GBE1 gene NM_000158: c.1571G>A: (p.Arg524Gln) mutation, while her parents and the younger brother carried a heterozygous mutation of c.1571G>A: (p.Arg524Gln). The mutation has been proved to be a rare mutation with frequency no greater than 0.01 in ExAC_ALL, ExAC_SAS and ExAC_EAS databases. The older brother was normal. The diagnosis of GSD IV was established and liver transplantation was recommended to her. The parents declined the liver transplantation. She died of liver failure at 22 months of age. This is the first homozygous mutation of GBE1 gene causing GSD IV being reported in mainland China. The identified mutations add to the list of GBE1 mutations of the hepatic form of GSD-IV.
Keywords
Glycogen storage disease type IV, Glycogen-branching enzyme 1 gene, Homozygous missense mutation.
Introduction
Glycogen storage disease type IV (GSD-IV; OMIM232500), also known as Andersen disease [1], is an autosomal recessive genetic disorder that results from a deficiency in the activity of the glycogen-branching enzyme GBE1, an amylo-(1,4-1,6)- transglycosylase (EC 2.4.1.18). The glycogen-branching enzyme adds short glycosyl chains in α-1,6 glycosidic links to the glycogen molecule to yield a branched polymer with increased water solubility and glycogen synthetic activity. The GBE1 deficiency leads to the storage of abnormal glycogen with fewer branch points and longer outer branches with low solubility in the liver, heart, and muscle [2-5]. Although GSDIV is a clinically heterogeneous disorder, its most common and classical form is progressive and hepatic. Here we describe a female infant with the classic hepatic presentation of GSD-IV and a homozygous known missense mutation in GBE1. This type of mutation of the homozygous form has not been reported in the hepatic form of GSD-IV, and this is the first case of GSD-IV confirmed by genetic analysis in mainland China.
Case Report
A 17-month-old Chinese girl was admitted to the Third Affiliated Hospital of Sun Yat-sen University (Guangzhou, China) in September 2015 with hepatosplenomegaly. She had shown signs of jaundice and failure to thrive since 15 months of age. The patient was the fourth child of healthy nonconsanguineous parents. Her older sister had died of liver failure at 12 months of age, while her two older brothers were healthy.
On physical examination, the biometric values were: body weight, 9 kg (10th percentile); body length, 75 cm (<10th percentile); and head circumference, 46 cm (50th percentile). She presented with mild jaundice of the skin and an enlarged liver and spleen (4 cm and 3 cm below the costal margin, respectively).
The initial laboratory evaluation revealed alanine transaminase, 135 U/L (normal, 3-35 U/L); aspartate aminotransferase, 759 U/L (normal, 13-35 U/L); an abnormal coagulation profile with elevated Prothrombin Time (PT) at 16.8 s (normal, 11.0-14.5 s) and Partial Thromboplastin Time (PTT) at 51.1 s (normal, 28.0-40.0 s); γ-glutamyl transpeptidase, high at 148 U/L (normal, 7-45 U/L); α-fetoprotein, 492.1 ng/ml (normal, ≤ 20 ng/ml); total bilirubin, 54.6 μmol/L (normal, 4.0-23.9 μmol/L); conjugated bilirubin, 41.4 μmol/L (normal, 0-6.8 μmol/L); total protein, 57 g/L (normal, 61-82 g/L); and albumin, 32 g/L (normal, 36-51 g/L). Fasting blood sugar, blood ammonia level, plasma lactic acid, uric acid, ceruloplasmin levels, creatine kinase, cholesterol and triglycerides, antinuclear antibodies, anti-smooth muscle antibodies, and liver/kidney microsome antibodies were in the normal range. Infectious diseases (toxoplasmosis, cytomegaly, herpes simplex, rubella, hepatitis B, and hepatitis C) were excluded. Metabolic investigations (tandem mass spectrometry and urine organic acid profile) were normal. Abdominal ultrasound revealed hepatosplenomegaly but no other abnormalities. Chest X-ray, echocardiogram, and electrocardiography revealed no abnormalities.
This study was approved by the ethical committee and institutional review board of the Third Affiliated Hospital of Sun Yat-sen University. Informed consent for the molecular genetic analysis was obtained from the parents. Peripheral blood samples from the patient and her healthy parents and two brothers were obtained. Genomic DNA was extracted from peripheral blood using the TIANamp blood DNA kit (Tiangen, Beijing, China) following the manufacturer’s instructions.
The generation of a targeted exome Illumina Library was performed according to the manufacturer’s protocol (MyGenostics, Beijing, China). A final library size of 350-400 bp including adapter sequences was selected. A total of 25 disease genes associated with glycogen storage disease (PGM1, LDHA, SLC37A4, PFKM, ALDOA, PHKB, G6PC, GYS1, GBE1, SLC2A2, PRKAG2, PHKA1, LAMP2, AGL, PYGM, GYS2, PYGL, PHKG2, ENO3, GAA, PRKAG3, GYG1, SLC17A3, PGAM2, PHKA2) were selected by a gene capture strategy using a GenCap Custom Enrichment Kit (MyGenostics) as previously described [6]. The enriched libraries were sequenced on an Illumina Solexa HiSeq 2000 sequencer for 100-paired reads. All of the exons and intronexon boundaries of the targeted genes were detected. After Solexa HiSeq 2000 sequencing, high-quality reads were retrieved from raw reads by filtering out the low quality reads and adaptor sequences using the Solexa QA package and the cutadapt program (http://code.google.com/p/cutadapt/), respectively. SOAPaligner program was then used to align the clean reads to the reference human genome (hg19). Subsequently, SOAPsnp, BWA, and GATK programs were used to retrieve and align variants and insertions or deletions (InDels). Variants and InDels were annotated using the MagicViewer to view the short read alignment and validate the candidate Variant and InDels. Suspicious mutations were validated by Sanger sequencing from all available family members. Nonsynonymous variants were evaluated by four algorithms-PolyPhen, SIFT, PANTHER, and PMUT to determine pathogenicity.
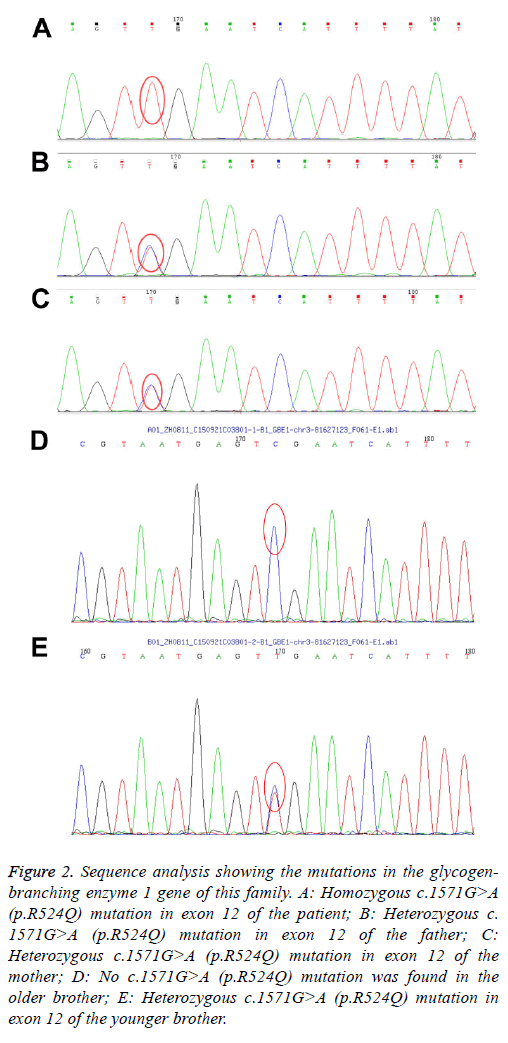
By the targeted exome sequencing of the sample, we obtained 194.41 Mb of effective sequences and mapped 127.63 Mb to the target regions. The average sequencing depth on the targeted regions was 225.46. More than 98.7% of the targeted regions were covered. The mean coverage of targeted exons for >10-fold was 91.7%, while that for >20-fold was 85.2%. GBE1 gene analysis revealed homozygosity for the G-to-A transition in exon 12 (c.1571G>A), leading to the replacement of an arginine by a glutamine at codon 524 (p.R524Q). Both parents and the younger brother were carriers of the mutation. The older brother is normal (Figures 1 and 2). After the diagnosis was confirmed by genetic analysis, liver transplantation was recommended. The parents declined the liver transplantation and discharged the patient from the hospital. Her clinical condition and liver function deteriorated rapidly thereafter and she died of liver failure at 22 months of age.
Figure 2: Sequence analysis showing the mutations in the glycogenbranching enzyme 1 gene of this family. A: Homozygous c.1571G>A (p.R524Q) mutation in exon 12 of the patient; B: Heterozygous c. 1571G>A (p.R524Q) mutation in exon 12 of the father; C: Heterozygous c.1571G>A (p.R524Q) mutation in exon 12 of the mother; D: No c.1571G>A (p.R524Q) mutation was found in the older brother; E: Heterozygous c.1571G>A (p.R524Q) mutation in exon 12 of the younger brother.
GSD is an inherited metabolic disorder of glycogen metabolism caused by the defects in glycogen synthesis or breakdown within muscles, liver, and other cell types due to lack of the enzymes required for these processes. There are over 12 types and they are classified based on the enzyme deficiency and the affected tissue. Liver and muscle have abundant quantities of glycogen and are the most common and seriously affected tissues. Those affecting the liver as the major organ are types I (glucose 6-phosphatase), III (debrancher), IV (brancher), VI (liver phosphorylase), and IX (phosphorylase b kinase) as well as glycogen synthase deficiency [7]. In this study, the patient presented with jaundice, hepatosplenomegaly, and failure to thrive; after the exclusion of infectious disease, autoimmune hepatitis, and some metabolic disease according to the results of tandem mass spectrometry and a urine organic acid profile, GSD was suspected. Although the histological findings in the liver were unknown because the parents denied a liver biopsy, the diagnosis of GSD-IV was confirmed by the clinical manifestations and mutation analysis findings. A homozygous missense mutation in the GBE1 gene was successfully identified via Capture Next-Generation Sequencing (CNGS) and validated by Sanger sequencing.
GSD-IV is an autosomal recessive genetic disorder due to a deficiency of the glycogen-branching enzyme GBE1. The gene of the enzyme is localized on chromosome 3p12.3. It has a coding sequence of 2,106 bp with 16 exons encoding a 702- amino acid protein (GBE) that is ubiquitously expressed [8]. The clinical presentation of GSD-IV is variable and involves the liver or the neuromuscular system [9]. The hepatic form is the most common presentation of GSD-IV and can be classified as classic progressive or non-progressive. The progressive form frequently presents in the first few months of life with hepatosplenomegaly, followed by progressive liver cirrhosis and death in early childhood. The non-progressive hepatic form is less common but can present in childhood with liver dysfunction, myopathy, and hypotonia. These individuals tend to survive without evidence of liver disease progression [10]. To date, more than 40 different mutations in the GBE1 gene have been described. Among them, 16 hepatic-form mutations have been reported [10-16]. The patient described here had a clinical course consistent with the classic hepatic form of GSD-IV and carried a homozygous previously described missense mutation c.1571G>A. Mutations in the GBE1 gene have been identified in all of the different clinical variants, and the same mutation has been identified in unrelated patients with different clinical presentations [9]. The missense mutation c.1571G>A was first reported in an infant with myopathy and non-progressive hepatopathy as a heterozygous form in a single allele [16]. It was later observed as a compound heterozygous mutation with c.38insA in the classic hepatic form [12]. However, the clinical features of homozygous c.1571G>A have never been reported. This is also the first case of GSD-IV confirmed by genetic analysis in mainland China.
We first reported the homozygous missense mutation c. 1571G>A of GBE1 gene in mainland China, the identified mutations add to the list of GBE1 mutations of the hepatic form of GSD-IV. Six of the 16 mutations reported in the hepatic form of GSD-IV are located in exon 12. Since exon 12 encodes part of the catalytic domain of the enzyme, it is a mutation hotspot to which we should pay more attention to [9].
Acknowledgements
We thank our patient’s parents for allowing us to publish this case. We also thank the staff members of this trial, our colleagues, and the study staff for their enormous efforts collecting the data and ensuring its accuracy and completeness. This study was supported by Guangdong science and technology project (2012B031800077).
References
- Andersen DH. Familial cirrhosis of the liver with storage of abnormal glycogen. Lab Invest 1956; 5: 11-20.
- Moses SW, Parvari R. The variable presentations of glycogen storage disease type IV: a review of clinical, enzymatic and molecular studies. Curr Mol Med 2002; 2: 177-188.
- Brown BI, Brown DH. Lack of an alpha-1,4-glucan: alpha-1,4-glucan 6-glycosyl transferase in a case of type IV glycogenosis. Proc Natl Acad Sci USA 1966; 56: 725-729.
- Servidei S, Dimauro S. Disorders of glycogen metabolism of muscle. Neurol Clin 1989; 7: 159-178.
- Lee YC, Chang CJ, Bai D, Chen YT, Yan YT. Glycogen-branching enzyme deficiency leads to abnormal cardiac development: novel insights into glycogen storage disease IV. Hum Mol Genet 2011; 20: 455-465.
- Huang XF, Xiang P, Chen J, Xing DJ, Huang N, Min Q, Gu F, Tong Y, Pang CP, Qu J, Jin ZB. Targeted exome sequencing identified novel USH2A mutations in Usher syndrome families. PLoS One 2013; 8: 63832.
- Matern D, Starzl TE, Arnaout W, Barnard J, Bynon JS, Dhawan A, Emond J, Haagsma EB, Hug G, Lachaux A, Smit GP, Chen YT.Liver transplantation for glycogen storage disease types I, III, and IV. Eur J Pediatr 1999; 158: 43-48.
- Nolte KW, Janecke AR, Vorgerd M, Weis J, Schroder JM. Congenital type IV glycogenosis: the spectrum of pleomorphic polyglucosan bodies in muscle, nerve, and spinal cord with two novel mutations in the GBE1 gene. Acta Neuropathol 2008; 116: 491-506.
- Assereto S, van Diggelen OP, Morava E, Cassandrini D, Carreira I, de Boode WP, Dilling J, Garcia P, Henrigues M, Rebelo O, ter Laak H, Minetti C, Bruno C. Null mutations and lethal congenital form of glycogen storage disease type IV. Biochem Biophys Res Commun 2007; 361: 445-450.
- Magoulas PL, EI-Hattab AW, Roy A, Bali DS, Fineqold MJ, Craigen WJ. Diffuse reticuloendothelial system involvement in type IV glycogen storage disease with a novel GBE1 mutation: a case report and review. Hum Pathol 2012; 43: 943-951.
- Li SC, Chen CM, Goldstein JL, Wu JY, Lemyre E, Burrow TA, Kang PB, Chen YT, Bali DS. Glycogen storage disease type IV: novel mutations and molecular characterization of a heterogeneous disorder. J Inherit Metab Dis 2010; 33: 83-90.
- Bruno C, van Diggelen OP, Cassandrini D, Gimpelev M, Giuffrè B, Donati MA, Introvini P, Alegria A, Assereto S, Morandi L, Mora M, Tonoli E, Mascelli S, Traverso M, Pasquini E, Bado M, Vilarinho L, van Noort G, Mosca F, DiMauro S, Zara F, Minetti C. Clinical and genetic heterogeneity of branching enzyme deficiency (glycogenosis type IV). Neurology 2004; 63: 1053-1058.
- Bao Y, Kishnani P, Wu JY, Chen YT. Hepatic and neuromuscular forms of glycogen storage disease type IV caused by mutations in the same glycogen-branching enzyme gene. J Clin Invest 1996; 97: 941-948.
- Shin YS. Glycogen storage disease: clinical, biochemical, and molecular heterogeneity. Semin Pediatr Neurol 2006: 13: 115-120.
- Said SM, Murphree MI, Mounajjed T, EI-Youssef M, Zhang L. A novel GBE1 gene variant in a child with glycogen storage disease type IV. Hum Pathol 2016; 54: 152-1526.
- Bruno C, DiRocco M, Lamba LD, Bado M, Marino C, Tsujino S, Shanske S, Stella G, Minetti C, van Diggelen OP, Dimauro S.A novel missense mutation in the glycogen branching enzyme gene in a child with myopathy and hepatopathy. Neuromuscul Disord 1999; 9: 403-407.