ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2016) Volume 27, Issue 2
Colistin resistance in Carbapenem-resistant Klebsiella pneumoniae strains.
| Fatma Kalem1*, Ayşe Gül Ergun1, Ömür Ertuğrul1, Serap Özçimen2, Hüsniye Şimşek3, Serap Süzük3,Özlem Ünaldı3, Rıza Durmaz3, Uğur Arslan4 1Microbiology Laboratory, Konya Numune Hospital, Turkey 2Clinical microbiology, Konya Numune Hospital, Turkey 3Department of Microbiology Reference Laboratory, Ministry of Health, Public Health Institution of Turkey, Ankara, Turkey 4Department of Microbiology, Selcuk University Selcuk Faculty of Medicine, Konya, Turkey |
| Corresponding Author: Fatma Kalem, Microbiology Laboratory Konya Numune Hospital Turkey |
| Accepted January 20, 2016 |
Objective: Because of the increase in the infections caused by carbapenem-resistant Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae ; nowadays colistin is used more frequently. In this study, the firstly detected colistin resistance in carbapenem-resistant KPC-producing K. pneumoniae strains were evaluated.
Material and methods: For identification and susceptibility testing; VITEK 2 Compact (bioMérieux, France) have been used. Because of resistance; MICs were studied with gradient test method in Microbiology Reference Laboratory, Public Health Institution of Turkey, Ministry of Health, Ankara, Turkey for confirmation. The presence of carbapenem resistance genes (OXA23, NDM1, OXA48, KPC, VIM ve IMP ) was investigated by Polymerase Chain Reaction (PCR) method. Pulsed Field Gel Electrophoresis (PFGE) method was used to determine the clonal relationships between strains. PCR and PFGE tests have been studied in Molecular Microbiology Research and Application Laboratory Department of Microbiology Reference Laboratories, Public Health Institution of Turkey, Ministry of Health, Ankara, Turkey.
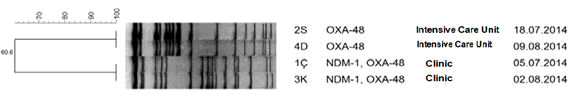
Results: All strains were resistance for carbapenems and colistin Two of four strains were isolated from patients hospitalized in intensive care and two of them were isolated from patients hospitalized in clinics. Resistance to carbapenems were confirmed genotypically. Two strains isolated from patients in clinics were positive for NDM1 and OX-48, and isolates from patients in intensive care unit were positive for only OXA-48 carbapenem genes. PFGE typing method described two clones that have a relationship with each other. The strains in which NDM1 and OXA-48 were together positive were in one clone and OXA-48-positive strains were in other clone.
Conclusion: The emergence of colistin resistant strains is a very important problem due to decrease of treatment options for infections caused by carbapenem-resistant KPC producing K. pneumoniae. Colistin should not be used alone, combination therapy should be preferred.
Keywords |
||||
| Colistin, Carbapenemase producing Klebsiella pneumoniae, PFGE | ||||
Introduction |
||||
| Enterobacteriaceae family which is one of the major factors of hospital and community-acquired infections causes increased morbidity and mortality especially with increased resistance rates [1]. Extended-spectrum beta-lactamases (ESBLs) lead to multidrug resistance by transfer between bacteria [2-5]. Carbapenems are important group of antibiotics used as a last option especially in ESBL producing multi-drug resistant Enterobacteriaceae. As a result of the increased use of antibiotics; Carbapenemases are widespread in Enterobacteriaceae family and particularly in K. pneumoniae so effective treatment options are decreasing [2,6]. Carbapenemases producing K. pneumoniae emerged in the late 1990s and has become a serious health problem in the world [7]. K. pneumonia which is a member of the intestinal flora is isolated as the causative agent in severe infections such as pneumonia, bacteremia, etc [8]. Carbapenemases producing K. pneumoniae strains are considered endemic in some areas. For example, studies by the European Antimicrobial Resistance Surveillance (EARS-Net) showed that the prevalence of carbapenem-resistant K. pneumoniae has increased from 1-2% to 15% in Italy between 2006-2009 [9]. Köseoglu et al. reported the rate of carbapenem resistant Enterobacteriaceae isolates as 11% in 2013 [10]. | ||||
| Due to this resistance spreading rapidly around the world; there have been a need for new therapeutic agents. Although because of side effects due to use of Polymyxin; this drug was out of use since early 1970s but that neurotoxic and nephrotoxic agent has become a preferable antimicrobial with increase of infections with resistant Enterobacteriaceae [4,11,12]. But excessive use of colistin led recently resistance to these drug [13]. Theresistance to polymyxin seen in K. pneumoniae strains is reported to be due to reduced affinity of colistin to lipopolysaccharide target [7]. In this study, colistin resistance in carbapenem resistant K. pneumoniae strains have been identified and these strains were evaluated. | ||||
Material and Methods |
||||
| In this study; VITEK 2 Compact (bioMérieux, France) were used for identification and antimicrobial susceptibility tests of K. pneumoniae strains isolated from different clinical samples sent to the Microbiology laboratory. Because of resistance to all drugs; tests were repeated. Then to verify the results; all of four strains were sent to the Department of Microbiology Reference Laboratory, Public Health Agency of Turkey. Reference laboratory MIC values of imipenem, meropenem, ertapenem, and colistin were studied with gradient test method (MIC Test Strips, Liofilchem, Italy). According to "Clinical and Laboratory Standards Institute (CLSI)" 2014 M100 S24; for imipenem and meropenem E-test results were considered as susceptible ≤ 1, 2 intermediate and ≥ 4 resistant, for ertapenem according to the same Standard ≤ 0.5 susceptible, 1 intermediate and ≥ 2 resistant and according to the European Committee on Antimicrobial Susceptibility Testing [EUCAST] 2014; colistin was considered as susceptible ≤ 2 and >2 resistant. The presence of carbapenem resistance genes (OXA23, OXA48, NDM1, KPC, VIM and IMP) were investigated by polymerase chain reaction(PCR) [14-17]. Primer sequences are given in Table 1. The multiplex PCR mixture was prepared by adding 10X PCR buffer (Fermentas, USA), 1.5 mm MgCl2, 200 nm from each primer, 200 mM each dNTP, 1.5 U Taq DNA polymerase (Fermentas, USA) and 2µl template DNA. Template DNA was obtained by the boiling method. For initial denaturation; after at 95°C for 5 minutes; processes were completed as denaturation at 95°C for 30 sec, connecting process at 58°C for 30 sec and 35 cycles extension process at 72°C for 1:30 sec and final extension at 72°C for 10 min. Amplification products were evaluated by agarose gel electrophoresis according to molecular weight standards. Pulsed Field Gel Electrophoresis(PFGE) method was used to determine the clonal relationship between the strains[18]. A 4 McFarland turbidity of bacterial suspension was mixed with low melting temperature agarose at a concentratio 1% (Bio- Rad Laboratories, Nazareth, Belgium). Blocks prepared from the mixture was incubated at 37°C for one hour in cell lysis solution (10 mm Tris-HCl (pH 7.2), 50 mm NaCl, 50 mm EDTA, 0.2% sodium deoxycholate, 0.5% sarkozil) and then was incubated in proteinase K solution (250 mm EDTA (pH 9.0), 50µg proteinase K and 1% sarkozil) at 50°C for one night. After the incubation; blocks were washed (each wash was at 50°C 30 minutes) 4 times in TE solution (10 mm Tris-HCl, 1 mm EDTA). Blocks were cut with 40 U XbaI enzymes. | ||||
| Electrophoresis was performed as two blocks. in 0.5 X TBE solution (44.5 mm Trizma base, 44.5 mm boric acid, 1 mm EDTA) in CHEF-DR II system (Bio-Rad Laboratories Ltd., Nazareth, Belgium). For first block; beginning and ending time was 1-30 seconds, pulse duration was 17 hours and for second block; beginning and ending time was 2.5 to 9 seconds, pulse duration was 6 hours. After electrophoresis; gel was stained with ethidium bromide(1µg/ml) and photographed with Gel Logic 2200 imaging system (Kodak Company, New York, USD). Gel DNA band profiles were analyzed with Gel Compar software (version 3.0; Applied Maths, Sint-Martens-Latem, Belgium). | ||||
| While DNA band comparing; tolerance was considered as 1.5% and optimization was considered as 1% and clonal relationships between the strains were evaluated according to Tenova criteria [19]. | ||||
Results |
||||
| In this study; K. pneumoniae strains were isolated from several clinical specimens of the patients in clinics and intensive care. All strains were resistant for imipenem, meropenem, ertapenem, and colistin by the automated system. Only one isolate was intermediate for meropenem. All tests were repeated. Because of same resistance profiles were observed; all strains were sent to Public Health Agency of Turkey, Department of Microbiology Reference Laboratory, the National Antimicrobial Resistance Surveillance Laboratory for verification. MIC values were similar with results determined by automated system. All strains were resistant for imipenem, meropenem, ertapenem, and colistin and one strain was detected intermediate for meropenem both with automated system and gradient test method. Also identifications were repeated with API and all strains were found as K. Pneumoniae. | ||||
| OXA-48 was detected in strains isolated from intensive care and NDM-1 and OXA-48 have been detected in strains isolated from clinics with PCR method in Molecular Microbiology Research Laboratory, Public Health Agency of Turkey. In table 1; Primer sequences and in Table 2; data on the strains have been presented. With PFGE typing method; two clones were described. PFGE results are shown in Figure 1. | ||||
DISCUSSION |
||||
| By the emergence of multidrug-resistant microorganisms; treatment options are decreasing. As an example; with the emergence and spread of carbapenemas producing K. pneumonia strains; outbreaks were seen in America, Europe and Asia and had become a major health problem [7]. Especially in America, Greece, and Italy; carbapenemproducing K. pneumonia strains have become common [6,7]. Infections caused by these strains cause high morbidity and mortality [20-22]. Therefore, treatment options, particularly in infections caused by K. pneumoniae producing carbapenemases decreased and tigecycline and colistin has become preferable. However, there are other treatment options, such as aminoglycoside particularly amikacin [2]. Excessive use of colistin to treat infections caused by resistant Enterobacteriaceae has led recently increased resistance to these drugs. The resistance rate of KPC-producing Klebsiella spp strains collected from health centers in North America, Latin America and Europe between 2000 and 2005 to polimiksin was reported as 6.7% [23]. In the same report; resistance was not detected for amikacin and tigecycline [23]. The increase of spread of multi-drug resistant strains is an important problem so measures should be taken [2]. | ||||
| In a retrospective study conducted in Greece showed that; of 150 patients with colonized with Gram-negative bacteria; 7 were colonized with colistin resistance K. pneumoniae strains [13]. | ||||
| In same region in a case-control study; the effect of factors such as age, sex, length of stay in acquisition of colistinresistant K. pneumoniae strains were investigated but no statistically significant effect was detected. Zarkoto et al. reported in their study that the main way for transmission of carbapenemase producing colistin resistant K. pneumoniae was horizontal transfer [24]. Colistin which is one of the few effective antimicrobials in infections caused by carbapenem resistant Acinetobacter spp and carbapenem producing Pseudomonas aeruginosa and K. pneumoniae is preferred recently [13,25]. Mostly use of colistin is the main risk factor for the emergence resistance to colistin [9,13]. The treatment of infections caused by carbapenemas producing K. pneumoniae is limited by the polymyxins and tigecycline, and especially if not used as combinations, they are not enough as therapeutic agent and leads to high mortality rates. Therefore it needs to be used in appropriate combinations. Tascini et al. investigated in vitro efficacy of tigecycline, imipenem, meropenem and colistin in different combinations in carbapenemase producing colistin resistance K. pneumoniae and colistin and rifampin combination was found to be the most effective combination [26]. In Italy three different clones for colistin resistant K. pneumoniae strains were detected. Although all the measures taken; spread could not be prevented [9]. Carbapenemase producing colistinresistant K. pneumoniae isolates have been reported from South Korea, too [27]. In the literature; the only report that we could achieved about colistin-resistant K. pneumoniae in Turkey has been reported from a private hospital [8]. In our study; according to genotypic analysis; two different clones were detected. Two strains isolated from clinics belonged to one clone and strains isolated from intensive care were belonged to other clone. The first colistin resistant strain was isolated from clinic for which imipenem and colistin were used for treatment. First K. pneumoniae strain isolated from this patient was sensitive to colistin. First colistin-resistant strain isolated in our hospital belongs to this patient. Then other resistant strains were isolated. This suggests that the horizontal spread of the resistant strains emerged with the use of colistin. Because of strict infection control policies implemented in our hospital; further spread has been prevented. Training about infection control and prevention is repeated periodically in our hospital. And also especially information has been given about this issue. Because of colistin use for the treatment of multiple drug resistance Gram-negative bacteria; the increase in ratio of colistin resistance is expected. Taking the necessary precautions to protect the latest treatment options and increase awareness about the use of antibiotics would be useful. | ||||
Tables at a glance |
||||
|
||||
Figures at a glance |
||||
|
||||
References |
||||
|