ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 18
Computational approach for diagnosis of malaria through classification of malaria parasite from microscopic image of blood smear
Niranjana Sampathila*, Nagaraja Shet and Akash Basu
Department of Biomedical Engineering, Manipal institute of Technology, Manipal Academy of Higher Education (MAHE), Manipal, Karnataka, India
- *Corresponding Author:
- Niranjana Sampathila
Department of Biomedical Engineering
Manipal institute of Technology
Manipal Academy of Higher Education (MAHE), India
Accepted on September 14, 2018
DOI: 10.4066/biomedicalresearch.29-18-970
Visit for more related articles at Biomedical ResearchMalaria is disease which is affecting millions of people and it is generally detected by examining the Red Blood Corpuscles (RBC) manually using microscope. However, the manual microscopic approach is time consuming, and lack of experts in the rural area, makes diagnosis of malaria very challenging one. The reported image processing approch extent the modern digital facilities to address the demand of automation, by developing a computerised facility for the detection of malaria using image processing technique. And this technological development could be a significant part of a modern digital telepathology. Proposed technology helps diagnose through the digital slide. Here the screening of microscopic images of a blood sample is achieved with color image processing approach that involves Red blood corpuscles (RBC) Segmentation, color space conversion, segmentation of the parasite, feature extraction and classification of malarial sample. The presented work detects plasmodium parasites from leishman stained microscopic blood images which in turn support pathologists for faster diagnosis. Neural network and rule based classifiers were used for the classification of blood images. The images belonging to malarial and non-malarial classes.
Keywords
Malaria, RBC, Color space, Plasmodium parasite, Neural network.
Introduction
Malaria is a public health problem and is a tropical disease which affects millions of people worldwide. The parasite named Plasmodium causes the life threatening disease malaria. In 2016, there were an estimated 216 million cases of malaria in 91 countries [1]. The female anopheles mosquito is responsible for the transmission of the disease. The malarial parasites multiply in the liver and infect Red Blood Corpuscles (RBC) present in the blood.
Malaria diagnosis involves parasite identification or antigens in the blood of a patient. The blood test can confirm the presence of malaria and its species. The tests are divided into microscopic tests and non-microscopic tests. Clinical diagnosis is dependent on symptoms of the patient and also based on the physical findings during examination. The initial symptoms of malaria include headache, chills, fever, sweat, muscle pain, vomiting and diarrhoea. In severe cases the clinical features may include anaemia, splenomegaly, hypoglycaemia, thrombocyte penia, renal dysfunction, and neurologic changes [2]. The symptoms are also found in some other diseases like flu and common viral infections. In severe malaria, clinical findings are prominent and the level of suspicion for malaria may increase. The Clinical findings must always be confirmed by a laboratory test [1,2].
Identification of malarial parasites in a blood sample using microscope is considered as a gold standard for laboratory confirmation of malaria. However, the malaria screening manually whole day is a tedious process. Even these analysis are time consuming and leads to inaccuracy and inconsistency in some cases. The computerised approach uses digitized blood slides that will be able to improve the consistence in diagnosis [3-9].
Reported the computerised automatic approach for detecting malarial parasite called Plasmodium vivax, by initially segmenting RBCs. The proposed algorithm is uses an image processing approaches for classification of images of the smear into normal and malarial. Thus it helps in upgrading the existing Lab technology into digital and even helps in automation.
Methodology
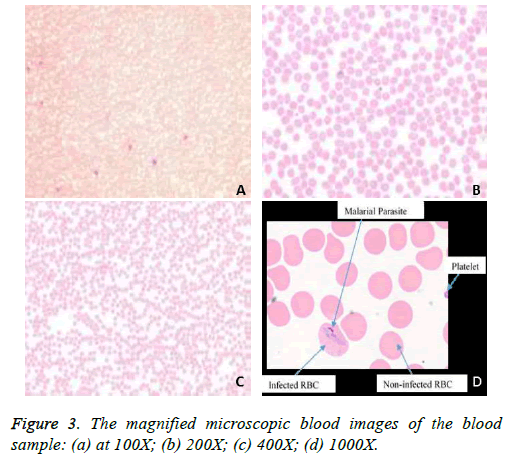
The Leishman stained thin blood smear slides were acquired with the fluorescence microscope, Olympus (BX51) at different magnification. The images recorded with the magnification of 1000X are indicates parasite clearly. The Olympus DP25 digital camera of 5 MP attached to the light microscope Olympus BX51, which is connected with the computer, along with the user interface software (DP2 BSW) are shown in Figure1. The acquired the blood image from the focused slide area are collected. The typical malarial thin blood smear image acquired at 40X magnification is shown in the Figure 2. Blood images acquired with the various magnifications such as 100X, 200X, 400X and 1000X are shown respectively in the Figures 3A-3d. A total of 143 images were considered for classification of malarial and non-malarial classes.
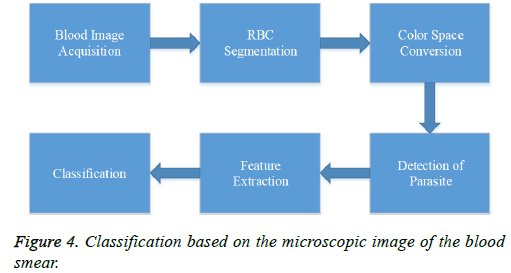
The image processing tool developed helps in the detection of malaria. The image is acquired from a thin blood slide using a microscope is processed to eliminate unwanted objects including platelets. The RBCs are segmented by thresholding the green channel of RGB image, later the malarial parasites are detected using saturation image of HSV colour space. The features are extracted from the gray scale image and parasite segmented image. The rule-based classifier and artificial neural network classifiers are used to classify the blood images into malarial or non-malarial. The approach for malarial identification using image processing technique is shown in the figure 4.
RBC segmentation
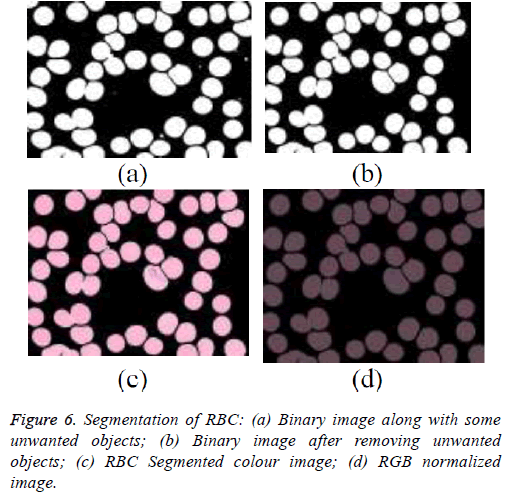
The acquired thin blood smear image has red blood corpuscles (RBC), malarial parasites, Platelets and other objects. But the proposed technique focus on diagnosis of malaria is based on examination of RBCs, since the malarial parasite infects the RBC. Thus the region of RBC is analysed with the Red, Green, Blue components of RGB image. The Green component of the RGB image has high contrast in order to distinguish background and foreground. In green layer of the RGB image the RBCs and the background are visible. With an appropriate threshold value RBC, the region of interest is extracted. Here foreground refers to the RBCs [3,5].
Then the binary image is super imposed with the original image so that the RGB image of extracted Red Blood Corpuscles in obtained. The normalized RGB image later converted into different color spaces, such as YIQ, YCbCr, CMY, LAB and HSV to identify the most suitable space for segmentation of malarial parasites.
Detection of parasite
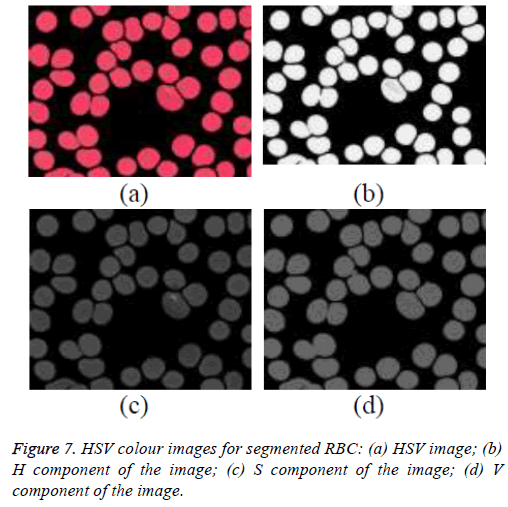
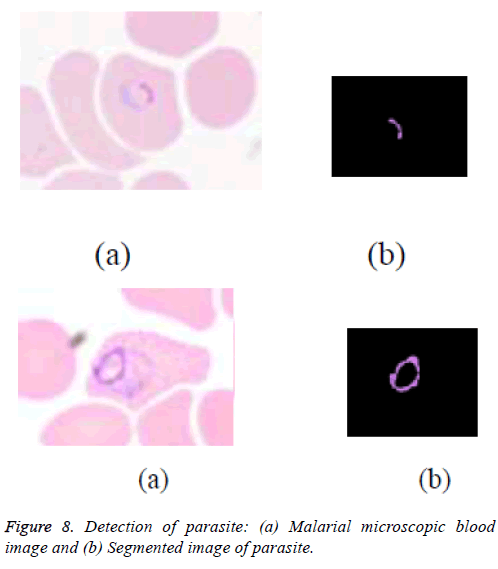
Studied the Hue-Saturation-Value (HSV) color space with the three components of HSV color space namely Hue, Saturation and Value and are analysed. Better signature of the parasite is seen in the saturation component of HSV image. The pixel values of parasite in saturation image are inspected. The parasite components have higher intensity value than the other objects in the image. Thus on the experimental basis, the threshold has been fixed to separate parasite from background.
But that resulted in the segmentation of RBC with the minor noise components which resemble the parasite. Hence morphological techniques are used to remove such minor unwanted objects.
Feature extraction
The classification of image involves the extraction of features and they include statistical, textural features, and geometrical features. The statistical features including mean, variance and standard deviation of a gray scale images are calculated. A gray level co–occurrence matrix (GLCM) is created from the grayscale image. Features are extracted from co-occurrence matrix to reduce space dimensionality. Geometrical feature such as area is calculated from a parasite segmented image [5].
Feature classification
The extracted features are used to classify the blood slides into non-malarial and malaria classes, using neural network classifier. Here the feed forward artificial neural networks are initially trained, the tested and evaluated. The training phase of an artificial neural network involves, providing the network with input feature vector and their respective target vectors (that is the desired outputs). The neural network classifier is designed for classification of blood images into malarial or non-malarial. The MATLAB is used for developing the proposed image processing approach, for identification of the malarial parasite from thin blood microscopic image.
Results
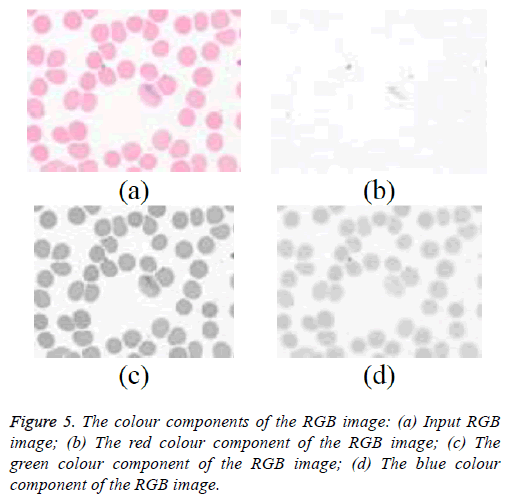
The microscopic blood image consisting of trophozoite stage Plasmodium vivax parasite is considered for the detection of malaria. The acquired images are in RGB image format and a typical blood image with Plasmodium vivax is shown in the Figure 5a. The red, green, and blue components that are extracted from the RGB image and are hown in the Figure 5b-5d. Each of the given color image components are analysed.
The green colour component has shown more details of RBC. That even gives a better contrast details between the background and foreground. Green component image is used for segmentation of the RBCs. The binary image obtained an appropriate threshold and given in the Figures 6a and 6b. This binary image generally contains platelet and other objects. The unwanted objects are removed using an area based threshold approach, the results are shown in Figure 6c. Because of the factors such as staining involved with blood slide the preparation and condition of the light during image acquisition still remind still challenging. To overcome this, the segmented images are normalised and the image is shown in the Figure 6d.
HSV colour space conversion
The different colour spaces are initially studied for the extraction of the parasite and finally the HSV colour space has shown better presence of parasite. The normalized RGB image is converted into HSV colour space and is shown in the Figure 7a. The hue, saturation and value components are extracted from the HSV image and are shown in the Figures 7b-7d respectively.
Detection of parasite
The saturation component of HSV colour space showed better contrast between background and the parasite region. Using a threshold technique on the saturation image, the parasite is segmented and is shown in the figure 8 (a). This image is then converted into binary and the noise components from the binary image are eliminated using morphological operations and its colour image obtained is shown in the figure 8 (b).
Classification of malarial parasite
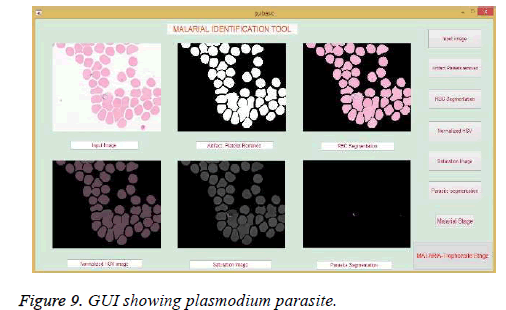
The classification involves classifying the given input image into malarial and nonmalarial. The GUI has been used to display the results of developed malaria identification algorithm. The GUI is developed to show the intermediate stages such as loading an image from a folder, RBC segmentation stage, parasite segmentation stage and finally classifying the parasite into non-malarial or malaria. The results obtained from the designed GUI are shown in the Figure 9.
The neural network classifier is fed with geometrical, statistical and textural features for the classification of microscopic blood images into malarial or non-malaria. The neural network is fed with the developed feature vector, which includes mean of the gray scale image, area of the segmented parasite, contrast, correlation, energy and homogeneity. The feature vector consisting of 6 features of 143 samples were used as an input to the neural network. The accuracies obtained for input feature vectors are tabulated in Table 1.
| Input data samples in percentage | Total Accuracy | ||
|---|---|---|---|
| Training | Validation | Testing | |
| 70 | 15 | 15 | 97.2 |
| 60 | 20 | 20 | 95.1 |
| 80 | 10 | 10 | 97.9 |
| Overall Accuracy | 96.7 | ||
Table 1. Accuracies obtained for classification of malarial and nonmalarial images using neural network.
Conclusion
The developed algorithm for identifying malarial parasites from the microscopic images of the blood sample has given interesting result. This computer based approach is faster and helps in consistent diagnosis. The developed algorithm removes the platelets and other smaller components from the microscopic blood image. The Red Blood Corpuscles (RBC) is segmented to detect parasite region. Among the different color spaces studied, the HSV color space is selected, for segmentation of RBC and parasite. The segmentation of parasite is performed using saturation image component in order to detect malarial class. The developed classification tool is promising with an average accuracy of 96.7%. The developed algorithm is a promising one could be used in the rural areas for screening the malarial patients or as part of digital tele-pathology application.
Acknowledgement
Authors would like to thanks the Haematology Lab, Kasturba Medical College (KMC), Manipal Academy of Higher Education (MAHE), Manipal providing the facility for collecting the data. Grateful thanks to Dr. Chethan Manohar, Professor, Department of Hemetology, K.M.C, MAHE, Manipal for valuable suggestions. Also they extend acknowledgements to the department of Biomedical Engineering, Manipal Institute of Technology (MIT), MAHE, Manipal.
References
- http://www.who.int/malaria/en/
- https://www.healthline.com/health/malaria
- Loddo A, Di Ruberto C, Kocher M. Recent advances of malaria parasites detection systems based on mathematical morphology. Sensors 2018; 18: 513.
- Das D, Mukherjee R, Chakraborty C. Computational microscopic imaging for malaria parasite detection: A systematic review. J Microscopy 2015; 260: 1-19.
- Shet N, Sampathila N. Detection of Plasmodium vivax from leishman stained malarial thin blood microscopic images. International Conference on Communication and Signal Processing, 2015, India.
- Jan Z, Khan A, Sajjad M, Muhammad K, Rho S, Mehmood I. A review on automated diagnosis of malaria parasite in microscopic blood smears images. Multimed Tools Appl 2017; 8: 9801-9826.
- Eshel Y, Houri-Yafin A, Benkuzari H. Evaluation of the parasight platform for malaria diagnosis. J Clin Microbiol 2017; 55: 768-775.
- Rakshit P, Bhowmik K. Detection of presence of parasites in human RBC in case of diagnosing malaria using image processing. IEEE Int Confer Image Informat Process 2013; 329: 9-11.
- Kurer DA, Gejji VP. Detection of malarial parasites in blood images. Int J Eng Sci Innovat Technol 2014; 3: 651-656.