ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2008) Volume 19, Issue 1
Discordance in the paraoxonase (Pon1)-192QR polymorphism
Department of Molecular Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia.
- *Corresponding Author:
- Rozaida Poh
Department of Molecular Medicine, Faculty of Medicine, University of Malaya
50603 Lembah Pantai, Kuala Lumpur, Malaysia.
Phone: +603 79674717
Fax: +603 79674957
e-mail: rozaiday@yahoo.com.
Accepted date: December 23 2007
Paraoxonase (PON1) has been implicated to have a cardioprotective role, due to its physical attachment with high-density lipoprotein. PON1192QR is a variation of the PON1 gene, the R allele being a risk factor for cardiovascular disease. Kinetic studies resulting in a plot of paraoxon versus diazoxon hydrolysis rates may be used to accurately predict PON1192 geno-type. In this study, paraoxonase and diazoxonase activities in plasma were measured spec-trophotometrically using plasma while PCR-based PON1192 genotyping was performed us-ing polymerase chain reaction followed by restriction digestion. The two-substrate assay-derived genotypes were cross-referred with those determined by PCR-based genotyping. When results did not concur, sequencing of the 99-bp region spanning codon 192 of PON1 was performed to verify the genotype. Concordant samples with high or low activities were sequenced for comparison. In addition, the rare PON1194WX polymorphism was examined as a source of discordance. The frequency of the PON1192Q allele in a Malaysian population comprising three ethnic groups (Malays, Chinese and Indians) was estimated to be 0.46, and that of the PON1192R allele, 0.54. Discordance between the genotype and phenotype was ob-served for two samples. One of the two subjects genotyped as PON1192QR and phenotyped as PON1192QQ. The sample showed low paraoxonase and diazoxonase activities. Sequencing confirmed that the genotype was PON1192QR. The other subject was genotyped as PON1192RR but phenotyped as PON1192QR. Sequencing showed that the genotype was in fact PON1192RR, with the subject showing relative high activities. The PON1194WX mutation was not detected in the sequenced samples and was not the source for discordance.
Keywords
PON1 gene, PCR-based genotyping, Two-substrate assay genotyping, Paraoxon, Diazoxon, Sequenc-ing.
Introduction
Paraoxonase1 (PON1) is an esterase belonging to a family of proteins that includes PON2 and PON3 [1]. The genes coding for the PON family are located on human chromo-some 7 (q21.22). PON1 has two exonic amino acid poly-morphisms, one at position 55 (methionine/leucine; M/L) and another at position 192 (arginine/glutamine; R/Q). PON1 is associated with high-density lipoprotein (HDL) particles, thus linked to the prevention of oxidative dam-age to low-density lipoprotein (LDL) particles. The pro-tective effect of HDL has been implicated in arresting the development of coronary heart disease (CHD), as well as other vascular complications of diabetes [2]. Several stud-ies describe the PON1192R allele as a significant risk factor for heart disease although a meta-analysis of 43 such studies have shown a weak association whereby the esti-mated relative risk of PON1192R for coronary heart disease was 1.12 [3]. Thus genotyping the diabetic individuals for both the PON1 192QR and 55LM polymorphisms updates available data and would be useful in identifying those individuals at the highest risk of developing complica-tions. The characteristics of PON1192QQ, -QR and -RR geno-types are such that the plasma activity differs towards the substrates paraoxon and diazoxon. The PON1192Q poly-morphism causes a ten-fold decrease in paraoxonase ac-tivity. The PON1192R mutation possibly reshapes the ac-tive site thereby improving the positioning of paraoxon [4]. In addition, hydrophobicity is common to most of PON1’s effective substrates, probably due to the hydro-phobicity and depth of PON1’s active site. Therefore, the poor and effective substrates bind at the active site with similar affinity, except that the mode of binding differs; the poor substrates would be inadequately positioned, resulting in a lower activity.
Material and Methods
Outpatients who attended the Diabetic Clinic, University of Malaya Medical Centre, Kuala Lumpur, and healthy volunteers residing in Selangor and Kuala Lumpur were screened for the study. This study was approved by the University of Malaya Medical Ethics Committee (MEC) and informed consent was obtained from all subjects.
Demographic characteristics of each subject were re-corded. All subjects were screened for the variations in the PON1 gene, including unmatched cases or controls. Venous blood samples were obtained by a trained phle-botomist under sterile conditions from subjects. Genomic DNA was extracted from whole EDTA blood using a Gene All Blood DNA Purification kit (General Biosys-tem, Seoul, South Korea). PON1 genotypes were deter-mined by the PCR-based methods [5]. To assess poly-morphism at position 192, the sense primer (5’ TAT TGT TGC TGT GGG ACC TGA G 3’) and the antisense primer (5’ CAC GCT AAA CCC AAA TAC ATC TC 3’) which encompass the codon 192 of the human PON1 gene were used. For the polymorphism at position 55, the sense primer (5’ AGA GGA TTC AGT CTT TGA GGA AA 3’) and the antisense primer (5’ CTG CCA GTC CTA GAA AAC GTT 3’) were used. PCR reactions were car-ried out using a Hybaid Omnigene thermal cycler (Hy-baid, Middlesex, UK). The hot-start PCR reaction mixture for the PON1-192 polymorphism contained 100 ng of DNA template, 0.5 μM of each primer, 1.5 mM MgCl2, 200 μM of each dNTP and 1 unit of Taq DNA poly-merase and hot-start antibody mixture (Biotherm PCR kit, Genecraft, Germany). The reaction mixture was denatured for 3 min at 93°C, followed by 35 cycles of 1 min of de-naturation at 93°C, 30 s of annealing at 61°C and 1 min of extension at 72°C. The final extension was 7 min at 72°C. The resulting 99 bp PCR product was then digested with 5 units of AlwI restriction endonuclease (New England Biolabs, Cambridge, MA, USA) overnight at 37°C, fol-lowed by size fractionation on 4% agarose gels with 10 μl matched undigested and digested samples loaded alter-nately into sample wells. Visualization was carried out with ethidium bromide staining. The R genotype (argin-ine) contains a unique AlwI restriction site which results in products of 63 and 36 bp, whereas the Q genotype (glutamine) is not cut.
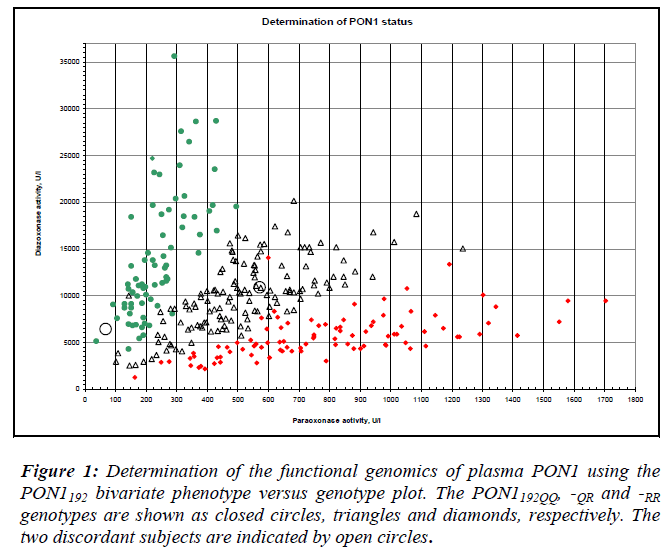
For the PON1-55 polymorphism, the PCR reaction mix-ture contained 100 ng of DNA template, 0.5 μM of each primer, 1.5 mM MgCl2, 200 μM of each dNTP and 1 unit of Taq DNA polymerase (Biotherm). After denaturation for 5 min at 94°C, the reaction mixture was subjected to 35 cycles of 30 sec of denaturation at 94°C, 30 sec of an-nealing at 61°C and 30 sec of extension at 72°C. The final extension time was 7 min at 72°C. The 386 bp PCR prod-uct was digested with NlaIII (New England Biolabs) in the presence of BSA overnight at 37°C. Digested products were separated in 3% agarose gels. The L genotype (leu-cine) does not contain an NlaIII site, whereas the M geno-type (methionine) contains an NlaIII site, giving rise to products of 296 and 90 bp [5]. For two-substrate assays, paraoxonase and diazoxonase activities of corresponding heparinised plasma samples were measured spectropho-tometrically (Varian Cary 50, Varian Inc. Scientific, Aus-tralia). A two-dimensional plot of diazoxon versus paraoxon hydrolysis rates allowed simultaneous PON1-192 genotyping and activity phenotyping of each individ-ual [6]. The genotypes obtained from both methods were compared. Sequencing was performed to validate geno-types in cases of discordance. Subjects with low activities for their PON1192 genotype were chosen for sequencing purposes as follows: PON1192QQ subjects with dia-zoxonase activities under 5000 U/L, and PON1192QR and –RR subjects with paraoxonase activities less than 200 U/L and 400 U/L, respectively. Reference to figure 1 shows that paraoxonase and diazoxonase activities are low for the subjects identified. The 99-bp PCR product was puri-fied by using a kit (MinElute, Qiagen, The Netherlands) and sequenced on an ABI 3730 DNA Analyzer (Applied Biosystem, California, USA). The sequenced products were compared using the nucleotide blast program against a nucleotide sequence database with reference to Genbank Accession #AF539592 [7]. The presence of the PON1194WX genotype was also examined based on the results from sequencing.
Results and Discussion
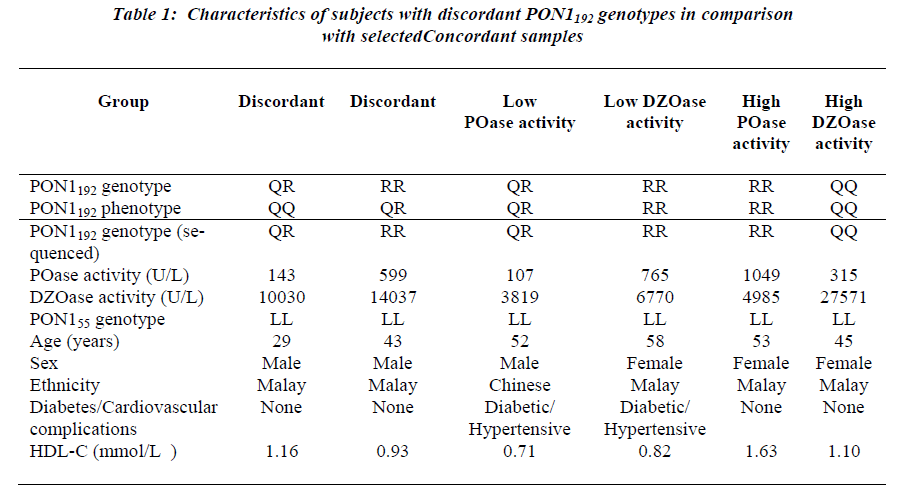
Examination of demographic characteristics showed that a greater proportion (60%) of the study population con-sisted of women. Malays constituted 47% of the study population, followed by Indians (33%) and Chinese (20%). The pooled age range was 17-97 years, with the mean age being 52 ± 13.5 years old. The subjects who were diabetic made up 52 % while the remaining 48 % were the non-diabetic controls. The observed frequencies of L, M, Q and R alleles in the pooled population were 0.912, 0.088, 0.459 and 0.541, respectively. Discordance between the genotype and phenotype was observed for two samples, 29- and 43-year-old, both healthy males. The 29-year- old subject was genotyped as PON1192QR and activity phenotyped as PON1192QQ. The sample had low paraoxonase and diazoxonase activities (Table 1). DNA sequencing confirmed that the genotype was PON1192QR. This subject had PON1 activity (<200 U/L) in the lowest tertile within its genotype group (Fig. 1). This low activity is consistent with the inactive allele hypothe-sis whereby the PON1192R allele is predicted to be inactive in this heterozygote.
The 43-year-old subject was genotyped as PON1192RR but phenotyped as PON1192QR. Sequencing showed that the genotype was PON1192RR, with the subject showing relat-ive high activities. For both samples, the presence of the rare PON1trp194stop polymorphism (PON1WX) which coded for the stop codon was assessed. All the selected samples were confirmed to be of the common PON1194WW geno-type. This subject showed comparatively high paraoxo-nase and diazoxonase activities. As the placement of acti-vity on the plot reflects the differential expression of the PON1192Q and PON1192R alleles, PON1192 homozygotes is genotype. In this case, the subject not only displayed dis-crepant genotypes despite being homozygous, it also dis-played high activities.
Repeated genotyping and phenotyping were performed to eliminate the possibility of error. In fact, other reports have noted the incidence of error when the salt-stimulation assay was used. Eckerson et al. [8] noted that one of the two individuals misclassified had low paraoxo-nase activity. The percentage of error was less than 1%. Browne et al. [9] reported a slightly higher error or analytical imprecision of 1.0-3.0%. These errors were re-solved when the genotypes were compared using various techniques. The rare PON1 protein truncation mutation PON1trp194stop may result in diminished PON1 activity. This alteration has been associated with the PON1192R allele (Jarvik et al., 2003). In the present study, all sub-jects sequenced for the mutation carried the PON1194WW genotype which was not the stop codon, thereby ruling out the effect of the stop codon on the loss of function of the particular allele. This is to be expected as the allele frequency of such a mutation occurring is relatively low at 1/458 PON1192R alleles in the population studied by Jarvik et al. (2003).
Acknowledgements
This study was supported by both the PPF FP030/2005D and PPP No P0117/2007A grants from the University of Malaya, Malaysia.
References
- Costa LG, Cole TB, Vitalone A & Furlong CE. Meas-urement of paraoxonase (PON1) status as a potential biomarker of susceptibility to organophosphate toxic-ity. Clin Chim Acta 2005; 352: 37-47.
- Mackness B, Durrington PN & Mackness MI. The paraoxonase gene family and coronary heart disease. Curr Opin Lipidol 2002; 13: 357-362.
- Wheeler JG, Keavney BD, Watkins H, Collins R & Danesh J. Four paraoxonase gene polymorphisms in 11 212 cases of coronary heart disease and 12 786 con-trols: meta-analysis of 43 studies. Lancet 2004; 363: 689-695.
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RBG, McCarthy A, Toker L, Silman I, Sussman JL & Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol 2004; 11: 412-419.
- Richter RJ, Jampsa RL, Jarvik GP, Costa LG & Fur-long CE. Determination of paraoxonase 1 status and genotypes. In Current Protocols in Toxicology. John Wiley & Sons, US 2004; pp 4.12.1-4.12.19.
- Jarvik GP, Jampsa R, Richter RJ, Carlson CS, Rieder MJ, Nickerson DA & Furlong CE. Novel paraoxonase (PON1) nonsense and missense mutations predicted by functional genomic assay of PON1 status. Pharmaco-genetics 2003; 13: 291-295.
- NCBI BLAST National Center for Biotechnology In-formation. 2003; www.ncbi.nlm.nih.gov/BLAST/.
- Eckerson HW, Wyte CM & La Du BN. The human serum paraoxonase/arylesterase polymorphism. Am J Hum Genet 1983; 35: 1126-1138.
- Browne RW, Koury ST, Marion S, Wilding G, Muti P & Trevisan M. Accuracy and biological variation of human serum paraoxonase 1 activity and polymorph-ism (Q192R) by kinetic enzyme assay. Clin Chem 2007; 53: 310-317.