ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2016) Volume 27, Issue 4
Effects of propofol on mRNA expressions of ion channels in Daphnia pulex
Laboratory of Anesthesiology and Critical Care Medicine, West China Hospital, Sichuan Province, 610041, PR China
- *Corresponding Author:
- Gui-zhi Du
Department of Anesthesiology and Critical Care Medicine
West China Hospital of Sichuan University
PR China
Accepted date: March 07, 2016
Propofol is multiple influences on organism function that could affect gene expressions of ion channels, cause arhythmia, impair brain cognitive functions, inhibit energy metabolism and increase hypoxic tolerance. To evaluate whether these phenomena might still exist in persistent low concentration of propofol exposure. We investigated changes in the mRNA levels of Daphnia 120 ion channel genes in response to exposed to persistent propofol (25 mM) for 240 min using quantitative PCR techniques. The heart rate (HR), glucose levels, phototactic behavior and hypoxic tolerance were recorded during the experiment. Sixteen of 120 ion channel transcripts in Daphnids were affected by propofol exposure: among them, 4 ion channel (1 Voltage-gated potassium (Kv) channel and 3 Ca2+-activated Cl- channels) genes were upregulated and 12 ion channel (3 Kv channels, 1 Cyclic nucleotide-gated channel, 1 Transient Receptor Potential Channel, 1 Glutamate-gated ion channel, 3-aminobutyric acid type A receptors, 1 DEG/ENaC channel, and 1 inositol 1, 4, 5-trisphosphate receptor) genes downregulated. Propofol did not show effect on gene expressions of Voltage-gated Calcium channels, Voltage-gated Sodium channels, Sodium-leak channel, Two-pore calcium Channel, AMPA receptor, NMDA receptor, Glutamate-gated chloride channels, Histamine-gated chloride channels, ATP-gated ion channels, Calcium release-activated calcium channel, Chloride intracellular channel, Bestrophin, Tweety channel and anoctamin. Additionally, propofol exposures not infuence HR, glucose levels, phototactic behaviors or hypoxic tolerances. Persistent propofol exposure even in small doses could affect multiple ion channels at mRNA levels. Systematical exploration of transcriptional changes of ion channels contributes to provide some insights into the understanding the mechanism of propofol.
Keywords
Propofol, mRNA, Ion Channel, Heart-beat, Glucose, Phototactic behavior, Hypoxic tolerance.
Introduction
Propofol is a short-acting sedative medication that is widely used in anesthesia, long-term sedation, and conscious sedation. Propofol could affect cell excitability by interacting directly with lots of ion channel proteins [1-4]. Long-term propofol use alther organism function and are linked to persistent changes in gene expression [5-7]. However, there is still no comprehensive data showing persistent low concentration of propofol exposure affected gene expressions of ion channels.
In general, previous studies just focused on the change in a single channel following propofol treatment. In fact, it is difficult to explain the pharmacological effects by propofol only with a certain channel mechanism. Drug-induced changes in gene expression play an important role in the pharmacodynamic response to drugs [5,6,8]. Changes in the expressions of ion channel genes might be provided some insights into the understanding the mechanism of propofol pharmacological effects.
Propofol is known to have multiple influences on organism function, including cause arhythmia [2,9], inhibit energy metabolism [10], impair cognitive functions [11] and increase hypoxic tolerance [12].
The effects of persistent propofol exposure in low concentrations on organism functions including heart rate (HR), glucose levels, phototactic behavior, hypoxic tolerance and the mRNA expressions of different classes of ion channels remain unknown. According to previous studies, we hypothesized that in a Daphnia pulex model, propofol could affect expression pattern of ion channel genes and organism functions.
Materials and Methods
Daphnia cultures and reagents
The method for culturing D. pulex was the same as previously described [13]. Briefly, the daphnids were kept in a temperature- and photocycle-controlled tank (20 ± 1°C, 16:8h light-dark cycle) and fed daily with a mixture of Saccharomycetes and Spirulina. Daphnia medium consisted of charcoal filtered tab water. One-fifth of the medium was renewed every second day. For all experiments, 3rd or 4th instars aged 8-12 days were used.
Since saltatory swimming style resembles the movements of Daphnia may complicate experimental results if the animals are immobilized by propofol while the animals in the control group are free to swim, we used 25 μm propofol in our experiment according to the finding of an EC50 of 58 μm for propofol immobilization of daphnids [14]. The Daphnia food was present during the experiment to avoid any complication from starvation of the animals. Daphnids were divided into two groups randomly: placebo group with saline exposure; propofol group with propofol exposure persistently for 240 min.
RNA extraction, reverse transcription and polymerase chain reaction (PCR)
At the end of the experiment, mRNA expressions of ion channels in two groups were evaluated. The tested ion channels in our study are divided into four groups based on their gating [15]: 1) volate-gated channels: voltage-gated potassium (Kv) channels, voltage-gated calcium (CaV) channels, voltage-gated sodium (NaV) channels, voltage-gated chlorine channels (ClC). 2) ligands- or second messengers-gated channels: two-pore calcium (TPC) channel, hyperpolarization-activated CNG (HCN) channels, cyclic nucleotide-gated (CNG) channels, glutamate-gated ion channel (iGluRs), nicotinic acetylcholine receptors (nAChRs), γ-aminobutyric acid (GABA) type A receptor (GABAAR), glutamate-gated chloride (GluCl) channels, histamine-gated chloride (HisCl) channels, ATPgated ion channels (P2X), calcium release-activated calcium (ORAI) channel, Inositol 1,4,5-trisphosphate receptor (IP3R), ryanodine receptor (RyR), bestrophin, tweety channels, anoctamin and calcium-activated chloride channels 3) chemical- or mechanical-gated channels: transient receptor potential (TRP) channels, ENaC/Deg ion channels, chloride intracellular channel (CLIC 4) other special channels: sodiumleak channel, nonselective (NALCN).
Total RNA extraction, reverse transcription and PCR were performed as previously described [13]. cDNA was generated using PrimeScriptTM RT reagent Kit DRR037A (TaKaRa), and amplified first by regular PCR to screen primers, which were designed using Primer 3 software based on the scaffold sequences (www.fleabase.org) [16]. Primers were used for quantitative PCR (qPCR) to quantify changes in gene expression after propofol treatment for 240 min.
Cycling parameters were 9°C for 30s to activate the DNA polymerase, then 40 cycles of 95°C for 5s, 55°C for 30s and 72°C for 30s. Melting-curves were performed to verify only a single product without primer-dimers. qPCR was carried out on an iQ5 system (Bio-Rad) using SYBR Premix Ex TaqTM II KIT DRR081A (TaKaRa). Among the 120 genes tested in our study, qPCR results were analyzed using the 2-ΔΔCT method and were normalized to β-actin [17].
Optical measurement of Daphnia HR and glucose assay
The heart beats of daphnids were video-recorded under an inverted microscope equipped with a digital eyepiece. The HR was counted offline using the software VitualDub-1.9.7 [18].
50 individual daphnids were homogenized with 500 μl deionized water, 100 μl of homogenate was used for glucose determination using Glucose assay Kit (GAGO-20, Sigma Aldrich), according to the manufacturer's instructions [18].
Phototactic assays
Phototactic behavior assays were based on a previous study conducted by Martins and colleagues [19]. The assay system consisted of 10 individuals placed into a glass test tube (20 cm height, 2.5 cm internal diameter) containing 70 ml of medium or a solution of 25 mM propofol in medium. This system was exposed to UV light from above using a portable 120V, 60W UV lamp, and phtototactic behavior of the daphnids was quantified by assessing the location of the individuals within the tube: Compartment I comprised the uppermost 14 cm of the tube and Compartment II comprised the bottom 2.5 cm. The data are presented as an index calculated by dividing the number of animals present in Compartment I by the total number of individuals in the assay system. Values obtained for the phototactic index ranged from 1 (all individuals present at Compartment I) to 0 (all individuals present at Compartment II). During the UV exposure period (240 min), the number of animals was measured at 20min intervals in three groups.
The HR, glucose and phototactic assays were recorded at 30, 60, 120, 180, 240 min after intervention.
Propofol treatment of Daphnia and survival test
The assessment of hypoxic tolerance was the same as previously described [18]. To quantify the survival rate, 10 individuals were placed in a 100 ml graduated cylinder. The survival rate was defined as the number of animals above the 50 ml mark over the total number animals tested. For hypoxic tolerance test, the choice of 4 mm sodium hyposulfite for hypoxic tolerance test was based on a series of measurements of the dissolved oxygen (DO) levels in different solutions from previous report [18]. The DO levels were measured by a dissolved oxygen meter (LIDA Dissolved 8, Shanghai, China). To quantify the survival rate, the daphnids were placed in a 100 ml graduated cylinder. The survival rate was defined as the number of animals above the 50-mL mark over animals was consecutively observed. This test started since propofol or saline exposure for 240 min in two groups. Survival rate of animals was observed at a 5-min interval during the period of hypoxic tolerance (60 min).
Statistical analysis
All data were presented as means ± SE with P < 0.05 as a criterion for significance. All data represented the average of 5 replicate experiments. HR and glucose levels were analyzed by one-way ANOVA, followed by Stuent-Newman-Keuls (SNK) tests when appropriate; phototactic index and survival rate with time were assessed by one-way ANOVA and Kruskal-Wallis H test, respectively.
All qPCR results were normalized to β-actin and then to control group. Differences in relative expression of genes were assessed using independent samples t test. Benjamini-hochberg multiple testing correction for false discovery rate (FDR) was applied and significance was set to FDR adjusted P-value <0.05 [20].
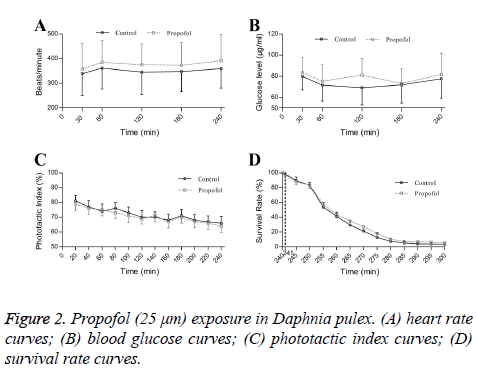
Propofol exposures did not affect heart rate (Figure 2A), blood glucose (Figure 2B) and phototactic behaviors of Daphnia during 240 min (Figure 2C). Propofol also not altered hypoxic tolerances in Daphnia pluex (Figure 2D).
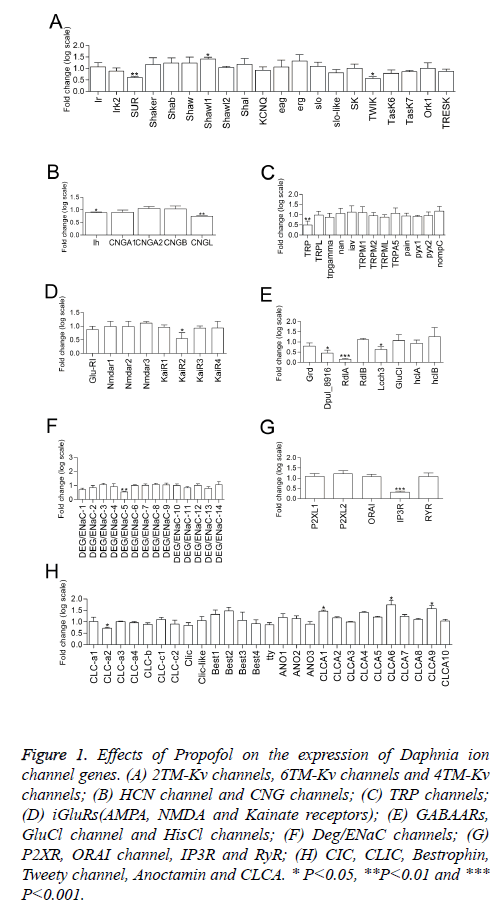
Figure 1: Effects of Propofol on the expression of Daphnia ion channel genes. (A) 2TM-Kv channels, 6TM-Kv channels and 4TM-Kv channels; (B) HCN channel and CNG channels; (C) TRP channels; (D) iGluRs(AMPA, NMDA and Kainate receptors); (E) GABAARs, GluCl channel and HisCl channels; (F) Deg/ENaC channels; (G) P2XR, ORAI channel, IP3R and RyR; (H) CIC, CLIC, Bestrophin, Tweety channel, Anoctamin and CLCA. * P<0.05, **P<0.01 and *** P<0.001.
Results
Statistical independent samples t test with Benjamini-hochberg multiple testing correction for false discovery rate (FDR) shows 16 transcripts of 120 ion channel gene were affected by propofol treatment (P<0.05). The transcription of 4 genes was upregulated by propofol treatment, including 1 Kv channel gene (the subunit of 6TM-Kv channel Shawl1) (Figure 1A) and 3 Ca2+-activated Cl- channel genes (CLCA1, CLCA6 and CLCA9)(Figure 1H). Meanwhile, propofol downregulated transcription of 12 genes, including 3 Kv channel genes (the subunit of 2TM-Kv channel SUR and the subunit of 4TM-Kv channel TWIK) (Figure 1A)1 HCN channel gene (Figure 1B), 1 CNG channel gene (CNGL) (Figure 1B), 1 TRP channel gene (homology of TRPC channel TRP) (Figure 1C), 1 iGluRs gene (the subunit of kainate receptor KaiR2) (Figure 1D), 3 GABAAR genes (Dpul_8916, RdlA and Lcch3) (Figure 1E), 1 DEG/ENaC channel genes (DEG/ENaC-5), 1 IP3R gene (Figure 1G) and 1 voltage-gated chlorine channel gene (CLC-a2) (Figure 1H). Propofol did not show effect on the expressions of CaV channels, NaV channels, NALCN channel, TPC channel, AMPA receptor, NMDA receptor, GluCl, HisCl, P2X channel, ORAI, CLIC, Bestrophin, Tweety channel and Anoctamin in the Daphnia genome at mRNA levels.
Discussion
Persistent propofol exposure in low concentration (25 mm) could affect gene expressions of ion channels. However, propofol exposures not influence heart rate, glucose levels, phototactic behaviors or hypoxic tolerances.
Sustained drug action could cause an increase in self-administration and thereby speed the user down the path to addiction and/or to overdose [5,6,21]. GABAA receptor play an important role in general anesthesia. Propofol at clinically relevant concentrations directly activates the GABAAR in the mammalian central neurons [1,22,23]. The mRNA profile changes may reflect the molecular targets not only in anesthetic actions, but also in chronic drug addiction [13].
Three genes of GABAA receptor subunits (Dpul_8916, RdlA and Lcch3) were downregulated by propofol in our results (Figure 1E). The transcription of GABAA receptor RdlA gene was also increased by midazolam and pentobarbital in our previous investigation [13]. The expression of different GABAA receptor subunits are affected by specific pharmacological manipulations [24].
The phenomenon of altering gene expression also existed in other ion channel subunits after propofol exposure. Arrhythmic substrates have been elucidated by ion-channel mRNA-expression profiling [7]. Propofol inhibits and slows activation of HCN channels at clinically relevant concentrations [9]. Propofol could slow the rate of HCN channels activation and rely on modulation of their gating to reduce heat rate in an isolated guinea pig heart [9]. In our findings, the gene expression of HCN channel was downregulated after propofol exposure (Figure 1B). Additionally, it is reported that propofol had an inhibitory effect on TRPC5 at ≥ 10 μm but not TRPM2 by patch-clamp recordings [3]. In our Daphids, 25 μm propofol could reduce the transcriptions of TRPC channel gene TRP and did not affect TRPM gene expression (Figure 1C). In Daphnia, propofol caused distinct mRNA expression profiles and this might provide insights into potential novel molecular targets involved in drug actions and addiction.
Pharmacological effects on ion channels function not always accompany the changes of gene expression in persistent low concentration of propofol exposure. Dose-dependent suppression of the various glutamente receptor channels were detected in response to propofol exposure in Xenupus oocytes [4]. The clinical concentrations (35 μm) of propofol suppress the NMDA receptor channels slightly [4]. Propofol downregulated gene transcription of Kainate receptor but showed no effect on AMPA receptor and NMDA receptor in our result (Figure 1D).
Clinically relevant concentrations of propofol could inhibit voltage-gated sodium channels-mediated Na+ influx, increasing intracellular [Na+] and releasing glutamate in synaptosomes [25]. The blocking action on cardiac L-type calcium channels of propofol shows marked resting block and use-dependent block [26]. Transient receptor potential (TRP) receptors TRPA1 and TRPV1 are key molecules for propofol-induced excitation of sensory neurons [27]. However, transcription of CaV channel genes, NaV channel genes, TRPA genes and TRPV genes were not altered by propofol in our experiment. Different propofol concentrations, exposure time and animals used might be ascribed for the difference.
Due to lack of the specific commercial antibody, we did not investigate the change of a responsive protein level with change in gene transcription. Despite the role of mRNA regulation is not explicitly implied in the theory, recent studies suggest that protein and mRNA expression levels are correlated [28,29].
Propofol exposure has been shown to reduce heat rate [2,9], increased glycogenolysis [10], caused cognitive defects [11] and provide neuroprotective effect in acute hypoxic injury [12]. However, propofol (25 μm) could not influence HR, glucose levels, phototactic behaviors and hypoxic tolerances in our research. Different propofol concentrations, exposure time and animals used might be ascribed for the difference.
In summary, systematical exploration of transcriptional changes of ion channels might greatly simplify the dissection of the core molecular pharmacological mechanism of propofol. Further experiments need conducting to link the changes in gene with function by propofol.
Grant Support & Financial Disclosures
National Natural Science Foundation of China [grant 81202607].
References
- Hara M, Kai Y, Ikemoto Y. Propofol activates GABAA receptor-chloride ionophore complex in dissociated hippocampal pyramidal neurons of the rat. Anesthesiology 1993; 79: 781-788.
- Ying SW, Abbas SY, Harrison NL, Goldstein PA. Propofol block of I(h) contributes to the suppression of neuronal excitability and rhythmic burst firing in thalamocortical neurons. Eur J Neurosci 2006; 23: 465-480.
- Bahnasi YM, Wright HM, Milligan CJ, Dedman AM, Zeng F, Hopkins PM, Bateson AN, Beech DJ. Modulation of TRPC5 cation channels by halothane, chloroform and propofol. Br J Pharmacol 2008; 153: 1505-1512.
- Yamakura T, Sakimura K, Shimoji K, Mishina M. Effects of propofol on various AMPA-, kainate- and NMDA-selective glutamate receptor channels expressed in Xenopus oocytes. Neurosci Lett 1995; 188: 187-190.
- Wu RS, Liu KC, Tang NY, Chung HK, Ip SW, Yang JS, Chung JG. cDNA microarray analysis of the gene expression of murine leukemia RAW 264.7 cells after exposure to propofol. Environ Toxicol 2013; 28: 471-478.
- Yoshida Y, Nakazato K, Takemori K, Kobayashi K, Sakamoto A. The influences of propofol and dexmedetomidine on circadian gene expression in rat brain. Brain Res Bull 2009; 79: 441-444.
- Nattel S, Frelin Y, Gaborit N, Louault C, Demolombe S. Ion-channel mRNA-expression profiling: Insights into cardiac remodeling and arrhythmic substrates. Journal of molecular and cellular cardiology 2010; 48: 96-105.
- Li Y, Liu YJ, Lv G, Zhang DL, Zhang L, Li D. Propofol protects against hydrogen peroxide-induced apoptosis in cardiac H9c2 cells is associated with the NF-kappaB activation and PUMA expression. Eur Rev Med Pharmacol Sci 2014; 18: 1517-1524.
- Cacheaux LP, Topf N, Tibbs GR, Schaefer UR, Levi R, Harrison NL, Abbott GW, PA. G. Impairment of hyperpolarization-activated, cyclic nucleotide-gated channel function by the intravenous general anesthetic propofol. J Pharmacol Exp Ther 2005; 315: 517-525.
- Acco A, Comar JF, Bracht A. Metabolic effects of propofol in the isolated perfused rat liver. Basic Clin Pharmacol Toxicol 2004; 95: 166-174.
- Sanou J, Goodall G, Capuron L, BourdalleBadie C, Maurette P. Cognitive sequelae of propofol anaesthesia. Neuroreport 1996; 7: 1130-1132.
- Harman F, Hasturk AE, Yaman M, Arca T, Kilinc K, Sargon MF, Kaptanoglu E. Neuroprotective effects of propofol, thiopental, etomidate, and midazolam in fetal rat brain in ischemia-reperfusion model. Childs Nerv Syst 2012; 28: 1055-1062.
- Dong C, Hu A, Ni Y, Zuo Y, Li GH. Effects of midazolam, pentobarbital and ketamine on the mRNA expression of ion channels in a model organism Daphnia pulex. BMC Anesthesiol 2013; 13: 32.
- Anmin. H, Changhong. D, Yunxia. Z, Guohua. L. Effects of Propofol, Etomidate and Ethanol on GPCR mRNA Expression in Daphnia pulex. Journal of Biomedical Engineering 2014; 31
- Alexander SP, Mathie A, Peters JA. Guide to Receptors and Channels (GRAC), 5th edition. Br J Pharmacol 2011; 164 Suppl 1: S1-324.
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132: 365-386.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and and the 2-ΔΔCT Method. Methods 2001; 25: 402-408.
- Sheng B, Liu J, Li GH. Metformin preconditioning protects Daphnia pulex from lethal hypoxic insult involving AMPK, HIF and mTOR signaling. Comp Biochem Physiol B Biochem Mol Biol 2012; 163: 51-58.
- Martins J, Soares ML, Saker ML, OlivaTeles L, Vasconcelos VM. Phototactic behavior in Daphnia magna Straus as an indicator of toxicants in the aquatic environment. Ecotoxicology and environmental safety 2007; 67: 417-422.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995; 289-300.
- Wang Y, Krishnan HR, Ghezzi A, Yin JC, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol 2007; 5: e265.
- Jayakar SS, Zhou X, Chiara DC, Dostalova Z, Savechenkov PY, Bruzik KS, Dailey WP, Miller KW, Eckenhoff RG, Cohen JB. Multiple propofol-binding sites in a gamma-aminobutyric acid type A receptor (GABAAR) identified using a photoreactive propofol analog. J Biol Chem 2014; 289: 27456-27468.
- Weerts EM, Ator NA, Griffiths RR. Comparison of the intravenous reinforcing effects of propofol and methohexital in baboons. Drug Alcohol Depend 1999; 57: 51-60.
- Uusi-Oukari M, Korpi ER. Regulation of GABAA receptor subunit expression by pharmacological agents. Pharmacol Rev 2010; 62: 97-135.
- Ratnakumari L, Hemmings HC. Effects of propofol on sodium channel-dependent sodium influx and glutamate release in rat cerebrocortical synaptosomes. Anesthesiology 1997; 86: 428-439.
- Yang CY, Wong CS, Yu CC, Luk HN, Lin CI. Propofol inhibits cardiac L-type calcium current in guinea pig ventricular myocytes. Anesthesiology 1996; 84(3):626-635.
- Fischer MJ, Leffler A, Niedermirtl F, Kistner K, Eberhardt M, Reeh PW, Nau C. The general anesthetic propofol excites nociceptors by activating TRPV1 and TRPA1 rather than GABAA receptors. J Biol Chem 2010; 285: 34781-34792.
- Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 2010; 6: 400.
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011; 473: 337-342.