ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2019) Volume 30, Issue 3
Efficacy of Lactobacillus ssp. in inhibiting the biofilm skin infections induced by Staphylococcus aureus pathogen
DOI: 10.35841/biomedicalresearch.30-17-390
Visit for more related articles at Biomedical ResearchBackground and aim: The escalating prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) has become a major public health threat. While lactobacilli were recently found useful in fighting various pathogens, limited data exist on their therapeutic potential for S. aureus infections. The aim of this study is to determine whether Lactobacillus Cell Free Supernatants (LCFS) were able to inhibit the biofilm produced by S. aureus. Material and methods: A total of 36 Staphylococcal bacteria were isolated from infected skin's patients. Antimicrobial susceptibility pattern of all tested isolates were determined using disk diffusion method. Biofilm production ability was detected using the Congo Red Agar (CRA) and the Microtitre Plate (MtP) assays. Furthermore, the efficacy of Lactobacillus CFS in inhibiting the biofilm skin infections induced by S. aureus isolates was estimated using MtP method. Results: Out of 36 S. aureus isolates, 34 (94.4%) and 20 (55.5%) showed positive results using CRA or MtP, respectively. All isolates showed high resistance against cefepime (97%), whereas imipenem was the most effective antibiotic against bacterial isolates. CFS exhibited significant anti-biofilm activities against the tested isolates. Conclusion: CFS of Lactobacillus spp. presents potential and safe alternative to synthetic antibiotics for inhibiting the biofilm skin infections induced by S. aureus.
Keywords
Biofilm, Lactobacillus, Staphylococcus aureus.
Introduction
Staphylococcus aureus is a gram-positive bacterium and the most pathogenic agent among all Staphylococcal species [1]. It is the causative agent of both simple skin infections and potentially life-threatening systemic complications such as toxic shock syndrome. The indiscriminate use of antibiotics for the treatment of bacterial infections induced by S. aureus has been suggested to be responsible for the appearance of Methicillin-Resistant S. aureus (MRSA) strains [2,3]. Production of biofilm facilitates persistence of S. aureus in the host tissue by protecting the bacterial cells against the mechanisms of the host defense [4,5]. Furthermore, it causes reduction of susceptibility to antibiotics, due to altered growth rate and delayed penetration of antimicrobial agents within the biofilm structure [6]. Considering the increasingly widespread ability of pathogens to generate persistent biofilm-related infections, an even more attractive proposal is to administer probiotics to prevent or counteract biofilm development [7]. Altered bowel flora is currently thought to play a role in a variety of disease conditions, and the use of Bifidobacterium spp. and Lactic Acid Bacteria (LAB) as probiotics has been demonstrated to be health-promoting, even if the success of their administration depends on the applied bacterial strain(s) and the targeted disease. LAB is useful for a variety of applications due to their therapeutic effects; these microorganisms are, in general, nonpathogenic, and thus have been assigned a “generally recognized as safe” status [8]. The aim of this study was to investigate the antibiofilm activity of lactobacillus sp. against skin infections induced by S. aureus isolates.
Materials and Methods
Patients and specimen collection
A total of 60 clinical specimens were randomly collected at the period from June to October 2014; from 80 patients (58 females and 22 males), aged below 12 y (2 to 11 y). Patients were recruited the Outpatient Clinics of Tanta University Hospitals, Egypt and suffered from skin lesion caused by diagnosed impetigo or burn infection. The study and consent form were approved by the local ethical committee (Approval code: 427/03/11). Specimens swabs were immediately placed in 2 ml phosphate- buffered saline (PBS; NaCl, 8 g/L; KCl, 0.2 g/L; Na2HPO4, 1.15 g/L; KH2PO4, 0.2 g/L) and were transferred to laboratory of Bacteriology in Botany Department, Faculty of Science, Tanta University, Tanta, Egypt.
Isolation and identification of bacteria
The collected specimens were plated to blood agar and nutrient agar. The resultant colonies were subcultured on mannitol salt agar (Oxoid, England) for preliminary selection of Staphylococcus aureus (yellow colonies) isolates. The recovered isolates were subjected to Gram reaction, catalase, coagulase, DNase tests for identification to the species level as described by Bergey’s Manual for Systematic Bacteriology [9].
Biofilm production by S. aureus isolates
Qualitative and quantitative phenotypic; Congo Red Agar (CRA) [10] and Microtiter Plate (MtP) [11] assays, respectively were used for detection of biofilm production by all tested isolates.
Congo red agar (CRA) assay
The isolates were cultured on CRA plates then incubated for 24 h at 37°C. The resultant colonies morphology classification adopting to a reference of six-color scale [12]. For each isolate the CRA plate test was repeated in triplicate, and consistent results were obtained.
Microtiter plate (MtP) assay
Overnight cultures were diluted 1:10 with Trypticase Soy Broth and 200 μL per well were seeded in 96-well microtiter plates. The plates were incubated at 37°C for 24 h. After four washes in phosphate buffered saline solution (pH 7.2), biofilms formed by adherent ‘sessile’ organisms in plate were fixed with sodium acetate (2%) and stained with crystal violet (0.1% w/v). Finally, the microtiter plates were rinsed under running tap water and the dye bound to the walls of the wells was resolubilized with 200 μL of 33% (v/v) glacial acetic acid per well. The absorbance or Optical Density (OD) was measured at 630 nm by using ELISA reader [13]. Strains were classified as Negative Producers (NP), Weak Biofilm-Producers (WP) and strong biofilm producing bacteria (SP). The mean OD value estimated from media control well was subtracted from all the test OD values. For each isolate the MtP test was repeated in triplicate.
Antimicrobial susceptibility testing
All the recovered S. aureus isolates were tested for methicillin resistance [14] on Muller Hinton agar using Oxacillin (OX) discs (1 μg). The isolates were considered MRSA if the zone of inhibition was 10 mm or less. Further, the antimicrobial susceptibility pattern of the tested S. aureus isolates to 18 antimicrobial agents was performed using; amoxicillin/ clavulanic acid (30 μg), cephlothin (30 μg), cefuroxime (30 μg), cefoperazone (75 μg), cefepime (30 μg), imipenem (10 μg), gentamicin (10 μg), rifampin (15 μg), levofloxacin (5 μg), lomefloxacine (10 μg), sulphamethoxazole/trimethoprim (25 μg), clindamycin (2 μg), erythromycin (15 μg), nitrofurantion (300 μg), vancomycin (30 μg), chloramphenicol (30 μg) and doxycycline (30 μg) by modified Kirby-Bauer single-disk diffusion technique on Muller Hinton agar containing 2% NaCl [15]. The results of the susceptibility tests were interpreted according to the criteria established by the Clinical and Laboratory Standards Institute [14]. Staphylococcus aureus ATCC 29213 kindly provided from culture collection of Faculty of Pharmacy, Tanta University, was used as reference strain.
Efficacy of different LCFS in inhibiting biofilm produced by the selected S. aureus isolates
The biofilm forming capability of the tested isolates in the presence of different LCFS were performed using MtP methods [13]. Lactobacillus plantarum subsp. plantarum DSMZ20174, Lactobacillus lactis subsp. cremaris, Lactobacillus rhamonsus ATCC 7469, Lactobacillus delbrueckii subsp. bulgaricus DMS20080, Lactobacillus acidophilus DSMZ20079T and Lactobacillus fermentum DSMZ20049 kindly provided from culture collection of Faculty of Science, Tanta University were used as references strains. Overnight Lactobacillus cultures contained 1.5 × 108 Colony Forming Units/ml (CFU/ml) were grown in MRS broth at 37°C for 24 h. Overnight cultures were centrifuged at 6000 rpm/min for 10 min at 4°C. The resulting supernatants were filtered through a 0.2 μm membrane filter. All supernatants were cultured on MRS agar in order to confirm the absence of lactobacillus cells. Different LCFS (200 μl) were added to each wells containing overnight culture of the tested S. aureus isolates followed by incubation for 24 h at 37°C, while control group was conducted without any additions of LCFS. The reference strains S. aureus ATCC was simultaneously used as positive control for biofilm production. Experiments were performed in triplicate.
Polymerase chain reaction (PCR) technique for detection of icaA gene
DNA was extracted from the selected staphylococcal isolates using bacteria DNA preparation kit (GeneJET™ Genomic DNA) according to manufacturer’s instructions. Amplification of the icaA gene was done using specific primers: 5́ TCTCTTGCAGGAGCAATCAA 3’ and 5́’ TCAGGCACTAACATCCAGCA 3’, yielding a PCR product of 188 base pairs (bp) [16]. These primers were designed from the published gene bank sequences (locus AF086783). The O ׳GeneRuler DNA Ladder Mix (100 bp or 1 Kbp) was used as a DNA size marker.
Determination of lethal dose 50 (LD50) of the tested S. aureus isolates
Two bacterial isolates; MSSA S15 and MRSA S12 representing different phenotypic biofilm production (strong biofilm producing isolate and weak biofilm producing isolate, respectively) were selected for determination of their LD50. Briefly, eight week old Swiss albino male mice weighing 18-20 g were obtained from the animal house at National Research Centre-Veterinary Division, Egypt. Mice were bred in the animal house, Faculty of Science, Tanta University. They were maintained under a 12 h light dark cycle at a temperature of 22 ± 2°C and fed with standard diet and water ad libitum. The study was conducted according to the ethical norms approved by the animal ethics committee guide lines of Tanta University. From overnight culture of the selected isolates, prepared bacterial cell suspension were introduced intraperitoneally (i.p) with different doses (109, 1010 or 1011 CFU/ml) to each group of mice. LD50 was determined as the concentration that caused 50% mortality rate of the injected mice [17].
Mortality rate=(No. of live mice/(No. of dead mice) × 100.
Statistical analysis
Data were compared using one way and two way (mean ± S.D) Analysis of Variance (ANOVA). Differences were considered significant when P ≤ 0.05.
Results
Biofilm production
In comparing of the biofilm production ability by the tested isolates (n=36) using CRA and MtP methods as disseminated in Table 1, it is clear that 27 (75%) and 9 (25%) of isolates were characterized as biofilm Producers (P). All results were compared with results that obtained by reference strain S. aureus ATCC 29213.
| Isolate code number | #CRA | *MtP | ||
|---|---|---|---|---|
| Production category | Production | (O.D) at 630 nm ± standard deviation | Production category | |
| S1 | +++ | P | 0.19 ± 0.05 | WP |
| S2 | +++ | P | 0.12 ± 0.01 | WP |
| S3 | +++ | P | 0.07 ± 0.01 | NP |
| S4 | +++ | P | 0.21 ± 0.02 | WP |
| S5 | ++ | P | 0.20 ± 0.06 | WP |
| S6 | +++ | P | 0.18 ± 0.01 | WP |
| S7 | ++ | P | 0.05 ± 0.04 | NP |
| S8 | + | WP | 0.41 ± 0.04 | P |
| S9 | ++ | P | 0.07 ± 0.01 | NP |
| S10 | +++ | P | 0.31 ± 0.09 | P |
| S11 | +++ | P | 0.13 ± 0.02 | WP |
| S12 | +++ | P | 0.15 ± 0.02 | WP |
| S13 | +++ | P | 0.18 ± 0.01 | WP |
| S14 | +++ | P | 0.08 ± 0.02 | NP |
| S15 | +++ | P | 0.44 ± 0.10 | P |
| S16 | +++ | P | 0.29 ± 0.01 | P |
| S17 | +++ | P | 0.30 ± 0.12 | P |
| S18 | + | WP | 0.09 ± 0.02 | NP |
| S19 | +++ | P | 0.08 ± 0.02 | NP |
| S20 | + | WP | 0.19 ± 0.01 | WP |
| S21 | +++ | P | 0.08 ± 0.02 | NP |
| S22 | +++ | P | 0.56 ± 0.10 | P |
| S23 | + | WP | 0.51 ± 0.08 | P |
| S24 | +++ | P | 0.06 ± 0.04 | NP |
| S25 | +++ | P | 0.15 ± 0.03 | WP |
| S26 | + | WP | 0.07 ± 0.01 | NP |
| S27 | + | WP | 0.06 ± 0.05 | NP |
| S28 | +++ | P | 0.06 ± 0.04 | NP |
| S29 | +++ | P | 0.36 ± 0.13 | P |
| S30 | + | WP | 0.15 ± 0.02 | WP |
| S31 | +++ | P | 0.06 ± 0.04 | NP |
| S32 | +++ | P | 0.27 ± 0.07 | P |
| S33 | - | NP | 0.11 ± 0.02 | WP |
| S34 | - | NP | 0.07 ± 0.01 | NP |
| S35 | ++ | P | 0.07 ± 0.01 | NP |
| S36 | ++ | P | 0.11 ± 0.02 | WP |
| S. aureus ATCC 29213 | +++ | P | 0.29 ± 0.01 | P |
| Control | 0.05± 0.01 | NP | ||
*CRA: Congo Red Agar method; P: Producer; WP: Weak Producer; NP: Non-Biofilm Producer. #MtP: Microtiter Plate Method; Producer (P)>0.240; Weak Producer (WP)>0.120 and <0.240; Non-biofilm Producer (NP)<0.120. S. aureus ATTC 29213 was highly producer (+++) appeared with black colonies.
Table 1. Biofilm production by different S. aureus isolates.
Antimicrobial susceptibility of the tested S. aureus isolates
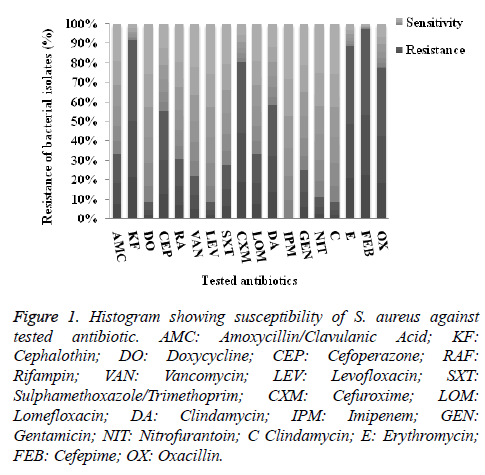
As illustrated in Figure 1, the incidence of antibiotic resistance of the investigated isolates showed that the highest resistance percentage were recorded against cefepime (97%), followed by cephalothin (92%), then erythromycin (89%), cefuroxime (83%) and oxacillin (77.8%). On contrary, among the tested drugs levofloxacin, chloromphenicol and tetracycline were the most active drugs where 8.3%, were resistant, in addition, all isolates were susceptible to imipenem.
Figure 1. Histogram showing susceptibility of S. aureus against tested antibiotic. AMC: Amoxycillin/Clavulanic Acid; KF: Cephalothin; DO: Doxycycline; CEP: Cefoperazone; RAF: Rifampin; VAN: Vancomycin; LEV: Levofloxacin; SXT: Sulphamethoxazole/Trimethoprim; CXM: Cefuroxime; LOM: Lomefloxacin; DA: Clindamycin; IPM: Imipenem; GEN: Gentamicin; NIT: Nitrofurantoin; C Clindamycin; E: Erythromycin; FEB: Cefepime; OX: Oxacillin.
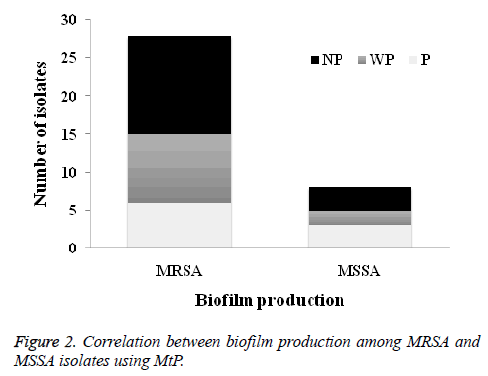
The results of OX susceptibility of these isolates confirmed the presence of 28 (77.8%) were MRSA and 8 (22.2%) were MSSA. Among MRSA tested isolates, 6 (21.4%) were biofilm produced while for MSSA, 3 (37.5%) were produced biofilms (Figure 2).
All isolates showed a high frequency of multiple [3-15] drug resistance, and up to 36.1% of the isolates showed multiple resistances to >9 antimicrobial drugs (Table 2). Noticeably high (72.2 and 83.3%) incidences of antibiotic resistance were detected among S. aureus S36 and S35, respectively.
| Isolate code | Antimicrobial resistance pattern | No. of resistance marker | Number of isolates exhibited the pattern | Resistance (%) | *Biofilm production using MtP |
|---|---|---|---|---|---|
| S20 | KF, FEB, OX | 3 | 1 | 16.7 | WP |
| S23 | CXM, FEB, OX | 3 | 1 | 16.7 | P |
| S34 | KF, E, FEB, OX | 4 | 1 | 83.3 | NP |
| S26 | KF, CXM, E, FEB, OX | 5 | 1 | 27.8 | NP |
| S10 | RAF, CXM, DA, E, FEB, OX | 6 | 1 | 33.3 | P |
| S21 | KF, CXM, DA, E, FEB, OX | 6 | 1 | 33.3 | NP |
| S29 | KF, LOM, DA, E, FEB, OX | 6 | 1 | 33.3 | P |
| S30 | KF, RAF, CXM , E, FEB, OX | 6 | 1 | 33.3 | WP |
| S33 | AMC, KF , CEP, CXM, E, FEB | 6 | 1 | 33.3 | WP |
| S1 | KF, CEP, SXT, CXM, LOM, E, FEB | 7 | 1 | 38.9 | WP |
| S2 | KF, CEP, CXM, LOM, E, FEB, OX | 7 | 1 | 38.9 | WP |
| S4 | KF, VAN, CXM, DA, E, FEB, OX | 7 | 1 | 38.9 | WP |
| S11 | KF, VAN, CXM, DA, GEN, E, FEB | 7 | 1 | 38.9 | WP |
| S13 | AMC, KF, CEP, CXM, DA, E, OX | 7 | 1 | 38.9 | WP |
| S14 | AMC, KF, CEP, CXM, E, FEB, OX | 7 | 1 | 38.9 | NP |
| S17 | KF, CEP, SXT, CXM, E, FEB, OX | 7 | 1 | 38.9 | P |
| S18 | KF, RAF, CXM, DA, E, FEB, OX | 7 | 1 | 38.9 | NP |
| S19 | KF, CEP, CXM, DA, E, FEB, OX | 7 | 1 | 38.9 | NP |
| S22 | KF, CEP, RAF, DA, E, FEB, OX | 7 | 1 | 38.9 | P |
| S27 | KF, CEP, RAF, CXM, DA, E, FEB | 7 | 1 | 38.9 | NP |
| S25 | KF, SXT, CXM, LOM, GEN, E, FEB, OX | 8 | 1 | 44.4 | WP |
| S7 | KF,CEP, RAF, CXM, LOM, DA, E, FEB, OX | 9 | 1 | 50 | NP |
| S8 | AMC, KF, CEP, VAN, CXM, DA, NIT , E, FEB | 9 | 1 | 50 | P |
| S31 | AMC, KF, CEP, VAN, SXT, LOM, DA, E, FEB | 9 | 1 | 50 | NP |
| S3 | AMC, KF,DO, CEP,SXT, CXM,C, E, FEB, OX | 10 | 1 | 55.6 | NP |
| S5 | AMC, KF, RAF, VAN, CXM, DA, NIT, E, FEB, OX | 10 | 1 | 55.6 | WP |
| S6 | AMC, KF, VAN, CXM, DA, GEN, NIT, E, FEB, OX | 10 | 1 | 55.6 | WP |
| S9 | KF, CEP, RAF, CXM, LOM, DA, C, E, FEB, OX | 10 | 1 | 55.6 | NP |
| S32 | KF, CEP, SXT, CXM, LOM, DA, GEN, E, FEB, OX | 10 | 1 | 55.6 | P |
| S12 | AMC, KF, CEP, VAN, CXM , DA,GEN, NIT, E, FEB, OX | 11 | 1 | 61.1 | WP |
| S24 | AMC, KF, CEP, LEV, SXT, CXM, LOM, GEN, E, FEB, OX | 11 | 1 | 61.1 | NP |
| S28 | AMC, KF, CEP, RAF, VAN, SXT, CXM, LOM, DA, GEN, E, FEB, OX | 13 | 1 | 72.2 | NP |
| S36 | KF, DO , CEP, RAF, LEV, SXT, CXM, LOM, DA, GEN, E, FEB, OX | 13 | 1 | 72.2 | WP |
| S35 | AMC, KF, DO, CEP, RAF, LEV, SXT, CXM, LOM, DA, GEN, C, E, FEB, OX | 15 | 1 | 83.3 | NP |
*MtP: Microtiter Plate Method; Producer (P)>0.240; Weak Producer (WP)>0.120 and <0.240; Non-Biofilm Producer (NP)<0.120. AMC: Amoxycillin/Clavulanic Acid; KF: Cephalothin; DO: Doxycycline; CEP: Cefoperazone; RAF: Rifampin; VAN: Vancomycin; LEV: Levofloxacin; SXT: Sulphamethoxazole/Trimethoprim; CXM: Cefuroxime; LOM: Lomefloxacin; DA: Clindamycin; IPM: Imipenem; GEN: Gentamicin; NIT: N
Table 2. Resistance pattern of S. aureus isolates.
Generally, in comparing antibiotic resistance and the detected biofilm by S. aureus clinical isolates, there is no obvious relation between biofilm production and resistance of isolates against antibiotics (Supplementary Data A).
Effect of LCFS on pre-formed biofilm of S. aureus
The highest biofilm producing MSSA (S8, S15 and S16) and MRSA (S10, S17, S22, S23, S29 and S32) isolates were selected for testing the influence of LCFS on their biofilm production, as shown in Table 3. Obtained results revealed that LCFS markedly affected produced or established biofilm of all investigated S. aureus isolates except CFS of Lactobacillus lactis that had insignificant effect on biofilm of S. aureus S10 isolate. LCFS were scattered or distributed pre-formed biofilm and reduced its intensity by converted them into planktonic mode. Statistical analysis showed a significant correlation between biofilm production and the isolates or LCFS at P=0.0001. Furthermore, the interaction between investigated S. aureus isolates and LCFS was significantly affected biofilm production at P=0.0001, as illustrated in Table 3.
| Isolate code | Biofilm production by S. aureus | ||||||
|---|---|---|---|---|---|---|---|
| Mean of (O.D) at 630 nm ± standard deviation (Biofilm category)# | |||||||
| Without addition of CFS (control) | With addition of CFS of standard strain of Lactobacillus | ||||||
| L. plantarum subsp. planturum DSMZ20174 | L. lactis subsp. cremaris | L. rhamonsus ATCC 7469 | L. delbrueckii subsp. bulgaricus DMS20080 | L. acidophillus DSMZ20079T | L. fermentum DSMZ20049 | ||
| MSSA S8 | 0.41 ± 0.04 (P) | 0.11 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.08 ± 0.0* (NP) |
| MRSA S10 | 0.31 ± 0.09 (P) | 0.11 ± 0.0* (NP) | 0.31 ± 0.0 ns (P) | 0.11 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.12 ± 0.0* (WP) | 0.17 ± 0.0* (WP) |
| MSSA S15 | 0.44 ± 0.1 (P) | 0.10 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.10 ± 0.0* (NP) | 0.18 ± 0.0* (WP) |
| MSSA S16 | 0.29 ± 0.01 (P) | 0.08 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.09 ± 0.0* (NP) | 0.06 ± 0.0* (NP) | 0.08 ± 0.0* (NP) |
| MRSA S17 | 0.30 ± 0.12 (P) | 0.08 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.12 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.06 ± 0.0* (NP) |
| MRSA S22 | 0.56 ± 0.10 (P) | 0.12 ± 0.0* (WP) | 0.10 ± 0.0* (NP) | 0.10 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.09 ± 0.0* (NP) | 0.09 ± 0.0* (NP) |
| MRSA S23 | 0.51 ± 0.08 (P) | 0.12 ± 0.0* (WP) | 0.06 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.06 ± 0.0* (NP) |
| MRSA S29 | 0.36 ± 0.13 (P) | 0.09 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.08 ± 0.0* (NP) | 0.08 ± 0.0* (NP) |
| MRSA S32 | 0.27 ± 0.07 (P) | 0.17 ± 0.0* (WP) | 0.09 ± 0.0* (NP) | 0.13 ± 0.0* (WP) | 0.08 ± 0.0* (NP) | 0.14 ± 0.0* (WP) | 0.19 ± 0.0* (WP) |
| S. aureus ATCC 29213 | 0.29 ± 0.01 (P) | 0.07 ± 0.0* (NP) | 0.11 ± 0.0* (NP) | 0.12 ± 0.0* (WP) | 0.09 ± 0.0* (NP) | 0.07 ± 0.0* (NP) | 0.08 ± 0.0* (NP) |
| Model effect (F-Mod) | 43.20*** | ||||||
| S. aureus isolates effect (F-Iso) | 15.04*** | ||||||
| CFS of Lactobacillus spp. effect (F-CFS) | 341.43*** | ||||||
| Isolates *CFS effect (F-Iso*CFS) | 10.61*** | ||||||
MRSA: Methicilin Resistance S. aureus, MSSA: Methicilin Sensitive S. aureus CFS: Cell Free Supernatant, Each reading was represented as mean ± standard deviations. #Biofilm category: Producer (P)>0.240; Weak Producer (WP)>0.120 and <0.240; Non-biofilm Producer (NP)<0.120. *Significant at P<0.05, ***highly significant P<0.001.
Table 3. Effect of CFS of different Lactobacillus spp. on biofilm producing S. aureus isolates using MtP.
Molecular detection of biofilm gene (icaA) encoded by the selected S. aureus isolates using polymerase chain reaction (PCR) technique
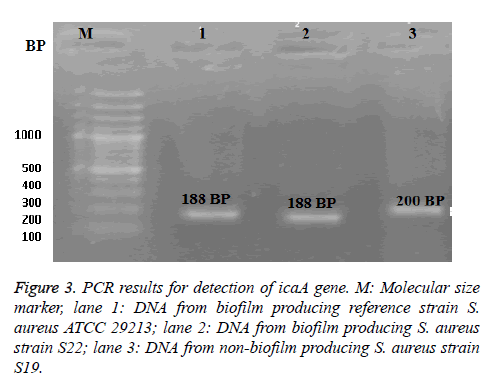
The PCR technique was applied to the selected two isolates; MRSA S22 and MRSA S19 (the highest and non bioflim producer, respectively) as shown in Figure 3. From the obtained results, the biofilm producing reference strain S. aureus ATCC 29213 was found to be harbor of icaA gene at 188 bp. It was noticed the presence of icaA gene in biofilm producing S22 isolate with identical band at 188-bp as well as standard strain. While non-biofilm producing S. aureus S19, icaA gene was found with a different molecular size band at 200 bp.
In vivo determination of LD50 by the tested S. aureus isolates
LD50 was detected by MSSA S15 and MRSA S12 (biofilm producer and non-biofilm producer, respectively) at 109 and 1010 CFU/ml, respectively after 24 h.
Discussion
Due to biofilm-associated staphylococcal infections are difficult to eradicate by routine antibiotics, this study aim to disrupt/scatter staphylococcal biofilm production using LCFS as an alternative routine. In our biofilm phenotype results, obtained data revealed that no correlation between the results obtained by CRA and that estimated by MtP, which is entirely consistent with previously reported data [18,19]. This could depend on the fact that biofilm production is affected by culture condition that causing a certain degree of variability in MtP results depending on the type of incubation medium and its production quantity. In such circumstances, some isolates can possibly be detected as non-producers, just because their phenotype is not completely expressed in basal TSB broth. For this reason, a number of improvements involving medium supplementation with sugars, salts, or ethanol have already been proposed by various authors in the attempt to favor phenotypic biofilm expression and its detection by MtP test [20].
Herein, the high prevalence of antibiotic resistance of the investigated S. aureus isolates was recorded. These results were comparable to the data obtained by Arsalan et al. [21] and Prakash [22]. Regarding to antimicrobial resistance pattern, bacterial isolates were found to be multiply resistant to 3-15 out of the 18 antimicrobials under test. These results support the finding by Ammendolia et al. [23]. Generally, it was noticed that no obvious relation between antibiotics resistance of all S. aureus clinical isolates and their biofilm production. In the same context, Ghasemmahdi et al. [24] documented that nearly all Salmonella typhimurium isolates revealed a high multiple antibiotic resistant with low biofilm producing capabilities which proposed low association between biofilm formation and antibiotic resistance. On contrary, Khan et al. [25] supported strong relation between antibiotics resistance and biofilm production by S. aureus isolates.
LCFS was able to disrupt/scatter biofilm of the tested S. aureus isolates and reduced its intensity via modifying the phenotypic expression (biofilm) to a planktonic mode of growth, which is entirely consistent with previously reported data [26,27]. Most probably LCFS was capable to control and disperse mature established biofilm of S. aureus with matrix degrading enzymes such as deoxyribonucleases, glycosidases and proteases, since DNA, proteins and Exoploysaccharides (EPSes) constituted the biofilm matrix [28,29]. Additionally, lacticin Q as bacteriocin produced from Lactobacillus lactis made stable pores on biofilm cells and was highly effective for the treatment of biofilm infections [30]. Some authors proved the important role of L. fermentum in prevention of skin wound in mice against both S. aureus and P. aeruginosa [31,32].
The ability of biofilm production by S. aureus also depends on the production of Polysaccharide Intercellular Adhesion (PIA) molecules, encoded by the intercellular adhesion (ica) locus including the icaA gene, icaB gene, icaC gene, and icaD gene. PCR amplification of the icaA gene demonstrates the inherent biofilm producing nature of the isolates [16]. Unexpectedly, our data reported that both biofilm producing and nonproducing clinical isolate was positive for icaA gene with different size 188 and 200 bp, respectively. This inactivation of icaA gene in non-biofilm producing isolate might be due to insertion mutation that changed the protein sequence and converted it to non-functional protein [32]. Thus, the absence of biofilm production in some staphylococcal isolates despite the presence of the ica operon might be due to the insertion of a 1332 bp sequence element, known as IS256 causing its inactivation [33]. Furthermore, Ziebuhr et al. [34] pointed out that the transposition of IS256 into the ica operon has been found to be a reversible process as after repeated passages of the Polysaccharide Intercellular Adhesion (PIA) negative insertional mutants, the biofilm-forming phenotype could be restored. In conclusion, nevertheless, irrespective of ica genes expression, ica-positive isolates should be considered to be potential biofilm producers [35].
Out of 36 S. aureus isolates, MSSA S15 and MRSA S12 were selected to in vivo determination of LD50, with aiming to detect role of biofilm on pathogenicity of S. aureus. It was found that S. aureus isolate with strong biofilm producing ability (MSSA S15) gave LD50 less than weak biofilm producer MRSA S12 isolate, which is entirely consistent with previously reported data [36]. Most probably biofilm ability increased severity of pathogenic S. aureus because it increased opportunity of pathogenic bacteria to adhere or attach to cell surface, colonize and finally invade cells resulted in infection [37].
With regards to relationship between antibiotic resistance and biofilm formation as a virulence factor, our study showed inverse relations where MSSA S15 was considered to be more virulent than MRSA S12. Thus means acquisition of antibiotic resistance in tested S. aureus isolates had been associated with the loss of pathogenic fitness and potential virulence factors. This phenomenon of inverse relationship between antibiotic resistance and virulence of our isolates has previously been reported [38-40], whose supports the hypothesis that, down regulation of virulence determinants by S. aureus might be a strategy to evade immune system detection, this perhaps will give the bacteria more time in acquiring mutations crucial to generate antibiotic resistance. Furthermore, multidrug resistant MRSA tended to harbor fewer virulence genes, whereas MSSA were more virulent but remained susceptible to many antibiotics.
Conclusions
Our observations indicate that LCFS is able to inhibit the preformed biofilm skin infections induced by Staphylococcus aureus isolate. Further studies to demonstrate the medical importance of Lactobacilli species as antibiofilm agent in clinical applications for skin infections are recommend.
Acknowledgement
The authors thank Dr Shereen F. Gheida, Department of Dermatology and Venereology, Faculty of Medicine, Tanta University, Egypt, for helping with the collection of samples from diagnosed cases.
References
- Lowy FD. Staphylococcal infections. Harrisons infectious disease. New York USA McGraw-Hill Professional 2010; 386.
- Rallapalli S, Verghese S, Verma RS. Validation of multiplesx PCR strategy for simultaneous detection and identification of methicillin resistant Staphylococcus aureus. Indian J Med Microbiol 2008; 26: 361-364.
- Grema HA, Geidam YA, Gadzama GB, Ameh JA, Suleiman A. Methicillin resistant Staphyloccus aureus (MRSA): a review. Adv Anim Vet Sci 2015; 3: 79-98.
- Jolivet-Gougeon A, Bonnaure-Mallet M. Biofilms as a mechanism of bacterial resistance. Drug Discov Today Technol 2014; 11: 49-56.
- Kang MS; Lim HS; Oh JS; Lim YJ; Wuertz-Kozak K; Harro JM; Shirtliff ME; Achermann Y. Antimicrobial activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathog Dis 2017; 75.
- Ramos AN, Sesto Cabral ME, Arena ME, Arrighi CF, Arroyo Aguilar AA ,Valdez1 JC. Compounds from Lactobacillus plantarum culture supernatants with potential pro-healing and anti-pathogenic properties in skin chronic wounds. Pharmaceut Biol 2015; 53: 350-358.
- Vuotto C, Longo F, Donelli G. Probiotics to counteract biofilm-associated infections: promising and conflicting data. Inter J Oral Sci 2014; 6: 189-194.
- Washington W, Allen S, Janda W, Koneman E, Procop G, Scheckenberger P, Woods G. Konemans Color atlas and text book of diagnostic microbiology (6th Edn.). JB Lippincott Williams and Wilkins Philadelphia 2006.
- Schleifer KH, Bell JA. Family VIII. Staphylococcaceae fam nov. Bergeys manual of systematic bacteriology (2nd Edn.) Springer-Verlag New York 2009; 3: 392.
- Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase-negative staphylococci. J Clin Pathol 1989; 42: 872-874.
- Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun1982; 37: 318-326.
- Arciola CR, Campoccia D, Gamberini S, CervelLati M, Dontati E, Montanaro L. Detection of slime production by means of an optimized Congo red agar plate based on a colourimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterials 2002; 23: 4233-4239.
- Uddin K ,Hossain T. Optimization of culture conditions for a biofilm forming bacterium. Inter J Pharma Bio Sci 2014; 1: 11-20.
- CLSI. Performance standards for antimicrobial susceptibility testing, twentieth informational supplement, CLSI document M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute 2010.
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966; 45: 493-496.
- Arciola CR, Baldassarri L, Montanaro L. Presence of ica A and ica D genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol 2001; 39: 2151-2156.
- Carlson E. Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect Immun 1983; 42: 285-292.
- Al-Mulla AF, Al-Khafaji ZM, Al-Kareemy KK. Comparison between methods for detection of biofilm produced by Staphylococcus epidermidis. Interl J Bio Pharmaceut Res 2013; 4: 1010-1014.
- Nasr RA, Abushady HM , Hussein HS. Biofilm formation and presence of icaAD gene in clinical isolates of Staphylococci. Egypt J Med Hum Genet 2012; 13: 269-274.
- Arciola CR, Campoccia D, Baldassarri L, Donati ME, Pirini V, Gamberini S. Detection of biofilm formation in Staphylococcus epidermidis from implant infections. Comparison of a PCR method that recognizes the presence of ica genes with two classic phenotypic methods. J Biomedic Mat Res Part A 2006; 76: 425-430.
- Arsalan A, Naqvi SB, Sabah A, Bano R, Ali SI. Resistance pattern of clinical isolates involved in surgical site infections. Pak J Pharmaceut Sci 2014; 27: 97-102.
- Prakash S. Carbapenem sensitivity profile amongst bacterial isolates from clinical specimens in Kanpur city. Indian J Crit Care Med 2006; 10: 250-253.
- Ammendolia MG, Di Rosa R, Montanaro L, Arciola CR, Baldassarri L. Slime production and expression of the slime-associated antigen by staphylococcal clinical isolates. J Clin Microbiol 1999; 37: 3235-3258.
- Ghasemmahdi H, Tajik H, Moradi M, Mardani K, Modaresi R, Badali A. Antibiotic resistance pattern and biofilm formation ability of clinically isolates Salmonella enterica serotype typhimurium. Inter J Enteric Pathog 2015; 3: 27372.
- Khan F, Shukla I, Rizvi M, Mansoor T, Sharma S. Detection of biofilm formation in Staphylococcus aureus. Does it have a role in treatment of MRSA infections? Trend Med Res 2011; 6: 116-124.
- Walencka E, Rolzalska S, Sadowska B, Rolzalska B. The influence of lactobacillus acidophilus-derived surfactants on staphylococcal adhesion and biofilm formation. Folia Microbiol 2008; 53: 61-66.
- Sambanthamoorthy K, Feng X, Patel R, Patel S, Paranavitana C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol 2014.
- Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 2010; 89: 205-218.
- McMillan A, Dell M, Zellar MP, Cribby S, Martz S, Hong E, Fu J, Abbas A, Dang T, Miller W, Reid G. Disruption of urogenital biofilms by lactobacilli. Colloids surf B: Biointerfaces 2011; 86: 58-64.
- Okuda K, Zendo T, Sugimoto S, Iwase T, Tajima A, Yamada S. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob Agents Chemother 2013; 57: 5572-5579.
- Valdez JC, Peral MC, Rachid M, Santana M, Perdigon G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: the potential use of probiotics in wound treatment. Clinic Microbiol Infect 2005; 11: 472-479.
- Sikorska H, Smoragiewicz W. Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Inter J Antimicrob Agents 2013; 42: 475-481.
- Kiem S, Oh WS, Peck KR, Lee NY, Lee JY, Song JH. Phase variation of biofilm formation in Staphylococcus aureus by IS256 insertion and its impact on the capacity adhering to polyurethane surface. J Korean Med Sci 2004; 19: 779-782.
- Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F ,Hacker J. A Novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence forcontrol of the polysaccharide intercellular adhesion synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol 1999; 32: 345-356.
- Oliveira A, Cunha Mde L. Comparison of methods for the detection of biofilm production in coagulase- negative Staphylococci. Oliveira and Cunha BMC Res Notes 2010; 3: 260.
- de Oliveira LF, Jorge AO, Dos Santos SS. In vitro minocycline activity on super infecting microorganisms isolated from chronic periodontitis patients. Braz Oral Res 2006; 20: 202-206.
- Franco JC, Julio C, Gonzalez V, Libertad, Gomez M, Sandra C. Virulence factors analysis of Staphylococcus aureus isolated from bovine mastitis in Mexico. e-Gnosis 2008; 6: 1-9.
- Gardete S, Kim C, Hartmann BM, Mwangi M, Roux CM, Dunman PM. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog 2012; 8: 1002505.
- Sapri HF, Sani NA, Neoh HM and Hussin S. Epidemiological study on Staphylococcus aureus isolates reveals inverse relationship between antibiotic resistance and virulence repertoire. Indian J Microbiol 2013; 53: 321-322.
- Jimenez JN, Ocampo AM, Vanegas JM, Rodríguez EA, Garces CG, Patino LA. Characterization of virulence genes in methicillin susceptible and resistant Staphylococcus aureus isolates from a pediatric population in a university hospital of Medellin, Colombia. Memorias do Instituto Oswaldo Cruz 2011; 106: 980-985.