ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 6
High glucose induces inflammatory reactions and changes in histonemodifying enzymes in rat mesangial cells
Keon-Cheol Lee1#, Jun Sik Lee2#, Sunhyo Jo2, DaeHee Kim3 and Sang Youb Han3,4*
1Department of Urology, Inje University Ilsan-Paik Hospital, Goyang, Korea
2Department of Life Science, BK21-plus Research Team for Bioactive Control Technology, College of Natural Sciences, Chosun University, Dong-gu, Gwangju, Korea
3Clinical Research Center, Inje University Ilsan-Paik Hospital, Goyang, Korea
4Division of Nephrology, Department of Internal Medicine, Inje University Ilsan-Paik Hospital, Goyang, Korea
#These authors contributed equally to this work
- *Corresponding Author:
- Sang Youb Han
Division of Nephrology
Department of Internal Medicine
Inje University Ilsan-Paik Hospital, Korea
E-mail: hansy@paik.ac.kr
Accepted date: December 11, 2017
DOI: 10.4066/biomedicalresearch.29-17-2049
Visit for more related articles at Biomedical ResearchHistone modifications, especially histone acetylation and methylation, are important for gene expression regulation in diabetic nephropathy. However, the general epigenetic changes in mesangial cells have not been characterized. We examined Histone Deacetylases (HDACs) and Histone Methyltransferases (HMTs), representative histone-modifying enzymes, during transient and persistent stimulation with High Glucose (HG) in Rat Mesangial Cells (RMCs). RMCs were incubated with 5.5 or 30 mmol/L Dglucose and were divided into 4 groups according to glucose stimulation over 24 h periods: group 1, no HG; group 2, initial HG followed by Low Glucose (LG); group 3, alternative HG and LG; group 4, persistent HG. The mRNA levels of interleukin (IL)-6 and IL1β were significantly higher in group 4 than group 1, indicating that transient and persistent exposure to HG could induce an inflammatory response. Both classes I (HDAC2, 3, 8) and II (HDAC4, 5) HDACs decreased as exposure to HG increased, and this decrease was particularly strong for HDAC2. However, class III HDAC levels did not differ among groups. Sirt1, 2, and 3 differed only slightly among groups. Mixed Lineage Leukaemia (MLL) 1, MLL2, and MLL3 were not affected by HG exposure, but other HMTs, e.g. G9a, Setdb1, and Suv39h1, showed a decrease. In conclusion, transient or persistent HG stimulation induced changes in inflammatory and histone-modifying deacetylase and methyltransferase in RMCs. These results suggested that HG decreased classes I and II HDACs and repressive HMTs, which could upregulates gene expression in RMCs.
Keywords
Diabetic nephropathy, Mesangial cell, Epigenetics, Histone deacetylases, Histone methyltransferases
Introduction
The pathogenesis of diabetic nephropathy is complex. Hyperglycaemia, advanced glycosylation end products, abnormalities in the polyol pathway, oxidative stress, and Transforming Growth Factor (TGF) are all involved in the progression of diabetic nephropathy [1]. Inflammation also contributes to the pathogenesis of diabetic nephropathy [2]. Infiltration of Monocyte Chemoattractant Peptide (MCP)-1, Interleukin (IL)-1, and adhesion molecule as well as macrophage accumulation in the kidney are correlated with albuminuria and renal fibrosis in diabetic nephropathy [3].
Epigenetic changes are heritable gene expression patterns not encoded in DNA. Histone modifications, especially histone acetylation and methylation, regulate gene transcription. Histone acetylation is regulated by two classes of enzymes, i.e. Histone Acetyltransferases (HAT), which acetylate specific lysine residues of histones, and Histone Deacetylases (HDACs), which remove acetyl groups from acetylated histones [4]. Histone methylation is regulated by Histone Methyltransferase (HMT). HAT-induced histone lysine acetylation leads to an open chromatin structure and thereby transcription factor and RNA polymerase II recruitment, whereas HDACs have the opposite function [5]. Histone methylation is also involved in posttranslational modifications, such as transcriptional regulation, and genome stability [5]. HMT regulates gene expression via methylation on a specific lysine residue. Generally, histone 3 lysine 4 methylation (H3K4me) is a transcriptional activation mark, and histone 3 lysine 9 methylation (H3K9me) is a repressive mark [6].
In addition, epigenetic mechanisms are associated with diabetes and diabetic complications [7,8]. In the Epidemiology of Diabetes Interventions and Complications (EDIC) study, observation study after the Diabetes Control and Complications Trial (DCCT), hyperglycaemia in the early study period had a prolonged beneficial effect on diabetic complications, such as myocardial infarction, stroke, and cardiovascular death, even though hyperglycaemia was controlled [9]. This metabolic memory effect can be explained by epigenetic changes. El-Osta et al. also showed that even transient High Glucose (HG) conditions induce persistent epigenetic changes [10]. Histone modification is directly or indirectly related to diabetic complications [10,11].
Emerging evidence suggests that diabetic nephropathy is extensively regulated by epigenetic modifications [12]. Changes in global histone modifications are correlated with progressive glomerulosclerosis in type 2 diabetic mice [13]. Although mice showed specific HG-induced epigenetic changes, the general epigenetic changes in mesangial cells have not been characterized. Accordingly, we examined representative histone-modifying enzymes, i.e. HDACs and HMT, in response to transient and persistent stimulation with HG in Rat Mesangial Cells (RMCs).
Materials and Methods
Rat mesangial cell culture and experimental design
RMCs were obtained by isolating glomeruli from the kidneys of male Sprague-Dawley rats by conventional sieving methods as previously described [14]. Briefly, RMCs were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Waltham, MA, USA) containing 20% foetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 44 mmol/L NaHCO3, and 14 mmol/L N-2-hydroxy-ethylpiperazine-N’-2-ethane sulfonic acid, at 37°C in a humidified atmosphere of 95% air and 5% CO2. Subconfluent cells were cultured in 0.1% foetal bovine serum for 24 h before the experiments. The cells were then incubated with 5.5 or 30 mmol/L D-glucose. The cells were divided into 4 groups according to the pattern of stimulation with HG every 24 h for 4 d as follows: group 1, no HG; group 2, initial HG followed by Low Glucose (LG); group 3, alternative HG and LG; group 4, persistent HG. Cells were cultured in triplicate and harvested at 96 h for the extraction of total RNA. All experiments were performed using cells between the 6th and 8th passages.
Histone-modifying enzymes
Representative molecules were selected to examine histone changes. Eight HDACs, including HDAC2, 3 and 8 for class I, HDAC4 and 5 for class IIA, and Sirt 1-3 for class III HDACs, were examined. Six HMTs were examined. H3K4me is typically associated with gene activation and can be mediated by several HMTs, such as SET1, MLL1-4 (mixed lineage leukaemia 1-4), and SET7/9. H3K9me, on the other hand, is generally associated with gene repression and can be mediated by SUV39H1 (Suppressor of variegation 3-9 homolog 1), G9a, and SETDB1/ESET (SET domain, bifurcated 1/ERGassociated protein with SET domain) [15,16]. MLL 1-3 were examined for H3K4me and G9a, Setdb1, and Suv39h1 were examined for H3K9me.
Inflammatory reactions
Inflammatory changes were evaluated based on the gene expression levels of Interleukin (IL)-6 and IL1β.
Reverse transcription-polymerase chain reaction (RTPCR)
Total RNA was extracted using TRIzol and reverse transcribed using a cDNA Synthesis Kit (Fermentas, Burlington, Canada) as previously described [17]. To synthesize cDNA, 0.5 μg of total RNA was primed with oligodT and reverse transcribed using a mixture of M-MLV RTase, dNTP, and reaction buffer (Promega, Madison, WI, USA). To measure the mRNA levels of inflammatory cytokines and epigenetic markers, primers were designed for target genes (Bioneer, Daejeon, Korea; Table 1). cDNA was amplified using the Gene Atlas G02 Gradient Thermal Cycler System (Astec, Fukuoka, Japan), e-Taq DNA Polymerase Kit (Solgent, Daejeon, Korea), and primers. PCR products were visualized using a fluorescent dye and a UV transilluminator.
| Target gene (product size) | Sequence | |
|---|---|---|
| HDAC-2 (249 bp) | Forward | 5’-TGGGCTGGAGGACTACATCA-3’ |
| Reverse | 3’-TCCAGCACCAATATCCCTCA-5’ | |
| HDAC-3 (227 bp) | Forward | 5’-ATGATGGCGACCATGACAAC-3’ |
| Reverse | 3’-TCAGGCCGTGAGAGTTTGAG5’ | |
| HDAC-4 (219 bp) | Forward | 5’-GCTTCCTTGTGGTGGTGTTG-3’ |
| Reverse | 3’-GCAACTGCCACGGAGTTAAA-5’ | |
| HDAC-5 (277 bp) | Forward | 5’-TCCTAGTCTCCGCTGGGTTT-3’ |
| Reverse | 3’-TTCTCTAGTGTGGCAACCGC-5’ | |
| HDAC-6 (269 bp) | Forward | 5’-GCTATGCGCAAAAGAAGCAC-3’ |
| Reverse | 3’GTGCAGGAAAGCAGCAATGT-5’ | |
| HDAC-8 (246 bp) | Forward | 5’-TCTGCGACTCCCTTGTGAAG-3’ |
| Reverse | 3’-ATATCCCTTCTGTGGCTGGG-5’ | |
| HDAC-9 (212 bp) | Forward | 5’-AGTCCCTTACTCAGGCGGAA-3’ |
| Reverse | 3’-AGTATGCGCTGCTGTGGAAC-5’ | |
| Sirt-1 (208 bp) | Forward | 5’-GACTTCCCAGACCTCCCAGA-3’ |
| Reverse | 3’-CTGCAACCTGCTCCAAGGTA-5’ | |
| Sirt-2 (240 bp) | Forward | 5’-TCTGCCACTACTTCATCCGC-3’ |
| Reverse | 3’-GGCTTTACCACGCTCTGACA-3’ | |
| Sirt-3 (204 bp) | Forward | 5’-GCCTGCAAGGTTCCTACTCC-3’ |
| Reverse | 3’-ACATCCCCTAGCTGGACCAC-5’ | |
| MLL-1 (282 bp) | Forward | 5’-GGTCATGCGTTCATCCTGAG-3’ |
| Reverse | 3’-AGTTGCTGGCAGGTATGCTG5’ | |
| MLL-2 (300 bp) | Forward | 5’-ATCACCGCCAATTTTAGCCT-3’ |
| Reverse | 3’-AGCATCCCTCAGTGCTCCTT-5’ | |
| MLL-3 (263 bp) | Forward | 5’-CTTGCTGGCTCGAGCTATTG-3’ |
| Reverse | 3’-ATATGCTGGGAAGAATGGGG-5’ | |
| Suv39h1 (229 bp) | Forward | 5’-TCCGCTACGATCTCTGCATC-3’ |
| Reverse | 3’-TTGCCATAATAAGCGGCATC-5’ | |
| G9a (208 bp) | Forward | 5’-GGTCCTTCTGAATGCCCAGT-3’ |
| Reverse | 3’-ATTGAGCTGCAGTGCAAACC-5’ | |
| Setdb1 (254 bp) | Forward | 5’-AGCATCAGCCTGAGGTCCTT-3’ |
| Reverse | 3’-GGCCTCCCTTGTGGTTATGT-5’ | |
| IL-6 (212 bp) | Forward | 5’-CCTTCCTACCCCAATTTCCA -3’ |
| Reverse | 3’-CGCACTAGGTTTGCCGAGTA-5’ | |
| IL-1β (287 bp) | Forward | 5’-GTGTCTTTCCCGTGGACCTT-3’ |
| Reverse | 3’-TCGTTGCTTGGTTCTCCTTG-5’ | |
Table 1. Primer sequence of target genes.
Quantitative real time reverse transcriptasepolymerase chain reaction (qRT-PCR)
The qRT-PCR was applied in the Applied Biosystems (StepOnePlus Real-Time PCR) from Thermo Fisher Scientific using the Power SYBRTM Green PCR Mater Mix kit. PCRgrade water was used as a negative control. GAPDH was used as the internal control to calculate the relative expression quantity of the objective genes in each sample, respectively.
Statistical analyses
We used non-parametric analysis because most of the variables were not normally distributed even after logarithmic transformation. The Mann-Whitney U test was used to compare differences between two groups. Statistical significance was defined as p value<0.05. All statistical analyses were performed using SPSS version 10.0 (SPSS Inc., Chicago, IL, USA). Data have been expressed as mean ± standard deviation.
Results
Inflammatory changes
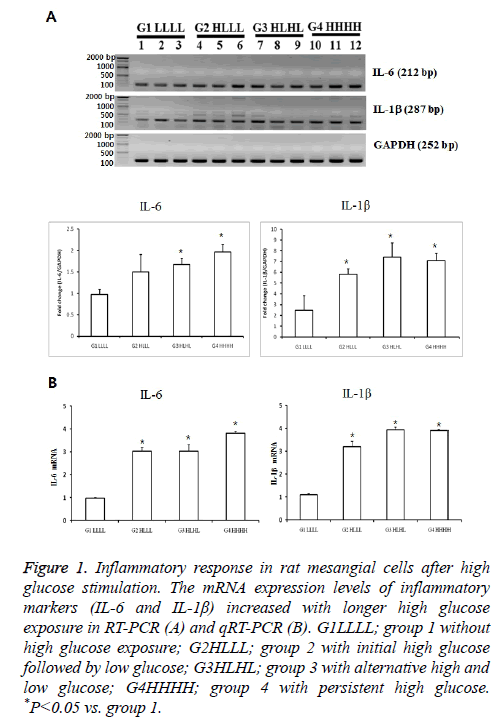
The mRNA expression of IL-6 was significantly higher in group 3 than in group 1 (p<0.05). The expression levels in group 4 tended to be higher than those in groups 2 and 3.The mRNA levels of IL1βwere higher in groups 2 and 3 than in group 1. IL1β levels in group 4 tended to be higher than those in group 2. We confirmed the significant results with real-time PCR again. These differences indicated that transient and persistent exposure to HG could induce an inflammatory response (Figure 1).
Figure 1: Inflammatory response in rat mesangial cells after high glucose stimulation. The mRNA expression levels of inflammatory markers (IL-6 and IL-1β) increased with longer high glucose exposure in RT-PCR (A) and qRT-PCR (B). G1LLLL; group 1 without high glucose exposure; G2HLLL; group 2 with initial high glucose followed by low glucose; G3HLHL; group 3 with alternative high and low glucose; G4HHHH; group 4 with persistent high glucose. *P<0.05 vs. group 1.
mRNA expression levels of histone deacetylases (HDACs)
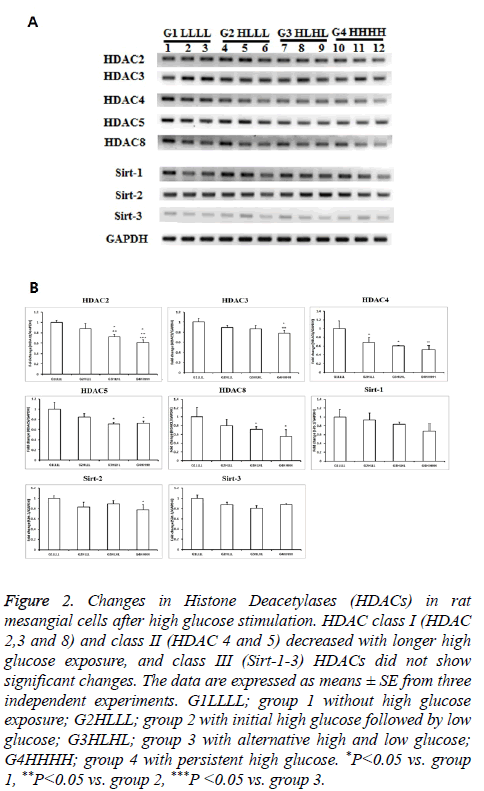
Classes I and II HDACs showed similar responses to HG. As exposure to HG increased, the levels of these HDACs decreased. Among class I and II HDACs, HDAC2 showed the most significant changes as exposure increased. HDAC2 levels were significantly lower in group 2 than in group 1, and were further decreased in group 4. HDAC3 expression only decreased in persistent HG conditions. The mRNA expression of HDAC4 decreased after transient HG exposure, and further decreased after persistent HG exposure. HDAC5 and HDAC8 expression levels were significantly lower in group 3 and group 4 than in other groups. All HDACs showed a decrease in response to transient or persistent HG exposure. However, class III HDACs were not affected by HG. Sirt1-3 showed only slight changes, even after persistent exposure to HG (Figure 2).
Figure 2: Changes in Histone Deacetylases (HDACs) in rat mesangial cells after high glucose stimulation. HDAC class I (HDAC 2,3 and 8) and class II (HDAC 4 and 5) decreased with longer high glucose exposure, and class III (Sirt-1-3) HDACs did not show significant changes. The data are expressed as means ± SE from three independent experiments. G1LLLL; group 1 without high glucose exposure; G2HLLL; group 2 with initial high glucose followed by low glucose; G3HLHL; group 3 with alternative high and low glucose; G4HHHH; group 4 with persistent high glucose. *P<0.05 vs. group 1, **P<0.05 vs. group 2, ***P <0.05 vs. group 3.
mRNA expression levels of histone methyltransferases (HMTs)
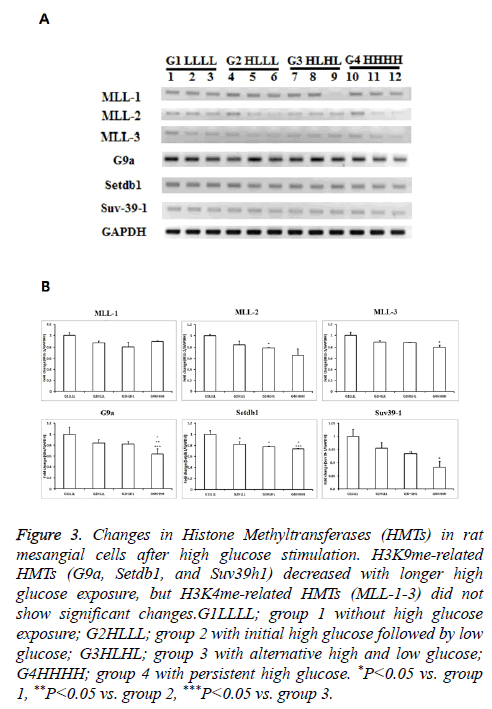
Although mRNA expression levels of MLL1-3 were not influenced by HG, the expression of other HMTs, such as G9a, Setdb1, and Suv39h1 decreased.
The mRNA expression levels of MLL1-3, which are related to H3K4me, did not differ among groups. The mRNA expression levels of G9a tended to be lower in group 3 than in group 1, and were significantly lower in group 4 than in groups 2 and 3. The mRNA expression levels of Setdb1 decreased significantly with a longer duration of HG exposure. Suv39h1 expression was significantly lower in group 4 than in group 1, and tended to be lower in group 4 than in groups 2 and 3. Considering that G9a, Setdb1, and Suv39h1 regulate H3K9me, HG influences lysine 9 methylation in RMCs (Figure 3).
Figure 3: Changes in Histone Methyltransferases (HMTs) in rat mesangial cells after high glucose stimulation. H3K9me-related HMTs (G9a, Setdb1, and Suv39h1) decreased with longer high glucose exposure, but H3K4me-related HMTs (MLL-1-3) did not show significant changes.G1LLLL; group 1 without high glucose exposure; G2HLLL; group 2 with initial high glucose followed by low glucose; G3HLHL; group 3 with alternative high and low glucose; G4HHHH; group 4 with persistent high glucose. *P<0.05 vs. group 1, **P<0.05 vs. group 2, ***P<0.05 vs. group 3.
Discussion
Transient or persistent HG stimulation induced changes in inflammatory genes and HDACs and HMTs in RMCs. Both classes I and II HDACs showed similar responses to HG, i.e. as HG exposure increased, the expression of classes I and II HDACs decreased. Their expression levels decreased further in response to sustained HG stimulation. Class III HDACs were not significantly influenced by HG.
With respect to HMTs, the levels of MLL1-3, which are related to H3K4me, were not affected by HG, but other HMTs, such as G9a, Setdbh1, and Suvh1, which are related to H3K9me, were downregulated. These results suggest that HG decreases class I and II HDACs and repressive HMTs, resulting in the upregulation of gene expression in RMCs.
Histone acetylation activates gene transcription in diabetic kidney models. HG induces genome-wide histone hyperacetylation in mesangial cells [18], including H3K9/14 hyperacetylation in approximately 5000 promoters of profibrotic genes, such as Tgfb1 and Ctgf, in glomerular mesangial cells [18]. These pro-fibrotic factors were further enhanced by an HDAC inhibitor, but were suppressed by an HAT inhibitor, confirming the importance of histone acetylation [18]. Increased PAI-1 and p21 expression levels are also associated with elevated promoter H3K9/14Ac levels in glomeruli of diabetic mice [19]. Conversely, the overexpression of HDAC1 and HDAC5 attenuates their expression [19].
However, contradictory results have been obtained. HDAC2, 4 and 5 were upregulated in streptozotocin-induced diabetic nephropathy, db/db mouse models, and diabetic patient kidney biopsies [20,21]. HDAC-2 mediated extracellular matrix accumulation and epithelial-to-mesenchymal transition in the diabetic kidney and in TGF-β1-treated epithelial cells [22,23]. HDAC inhibitors, including Trichostatin A (TSA), have been reported to attenuate diabetic nephropathy [24-26]. Yoshikawa et al. also reported that an HDAC inhibitor suppressed TGF-1β-induced epithelial-to-mesenchymal transition in human renal proximal tubular epithelial cells [27]. An HDAC inhibitor did not directly affect TGF-1β, but TSA inhibited TGF-β-induced epithelial-to-mesenchymal transition. This effect of the HDAC inhibitor was explained by the upregulation of histone acetylation by inhibitors of DNA binding/differentiation 2 and bone morphologic protein 7 [27]. In our study, HG reduced the expression of classes I and II HDACs, but not class III HDACs, in RMCs. Therefore, genespecific histone modification is important, even though histone lysine acetylation is generally associated with gene activation and HDACs have opposing functions.
Recently, HDAC inhibitors have been identified as promising drugs for the treatment of several diseases, including diabetic nephropathy. However, several reports have indicated that epigenetic regulation, including the effects of HDAC inhibitors, could be cell- and gene-specific. Initially, Cai et al. reported that glucose stimulates osteopontin expression and this expression is inhibited by an HMT inhibitor, but not by an HDAC inhibitor. The HDAC inhibitor even increased the expression of Spp1, an osteopontin gene [28]. Second, TSA, in general, can induce histone hyperacetylation to increase gene expression. However, TGF-β1 target genes vary. TSA inhibited TGF-1-induced fibronectin and Col1a2 by inhibition of Sp1 recruitment at the Col1a2 promoter. However, it increased Ecadherin by increasing histone acetylation [26]. These differences suggested that the effects of HDACs are genespecific, and both histone and non-histone-dependent mechanisms are involved. Third, the effects of renoprotective drugs are histone-modifying enzyme-specific. Reddy et al. reported that losartan inhibits H3K9/14Ac and H3K36me3 at receptor for advanced glycation end products (RAGE) and plasminogen activator inhibitor-1 (PAI-1), respectively. However, losartan does not change the decreased H3K9me2, H3K9me3, and H3K27me3 marks at RAGE and PAI-1 genes. Additionally, losartan dose not influence histone changes at MCP-1, which is associated with the pathogenesis of diabetic nephropathy [29].
Histone changes relating to histone methylation are very complicated. Gene expression depends on the type and location of lysine methylation. Up-regulation of H3K4me1/2/3 marks contributes to increased gene expression in diabetic nephropathy [30-32]. Inflammatory gene activation is also related to a loss of repressive marks, such as H3K9me, in inflammatory genes, such as IL-6 and MCP-1, in diabetic vascular smooth muscle cells. This loss of repressive H3K9me3 in db/db vascular smooth muscle cells is associated with decreased protein levels of Suv39h1, a known HMT that mediates H3K9me3, and is associated with repressed states of chromatin [33]. In our study, H3K4me-related HMTs, such as MLL1-3, were not influenced by HG. However, H3k9merelated HMTs, such as G9a, Setdb1, and Suv39h1, were significantly downregulated in HG-induced RMCs, even after early transient HG exposure, which could decrease repressive marks, e.g. H3K9me.
Various efforts to block inflammatory reactions have not been effective in diabetic nephropathy. The control of epigenetic changes is a promising option to manage diabetic nephropathy. In diabetes, oxidized low-density lipoprotein and endoplasmic reticulum increase the protein levels of SET7, a H3K4me HMT, and promote its nuclear translocation and enrichment at profibrotic gene promoters [32,34]. The overexpression of Suv39h1 in vascular smooth muscle cells partially reversed inflammatory gene expression in diabetic db/db mice [33]. Our results showed that changes in inflammatory cytokines corresponded to the downregulation of class I and II HDACs and repressive HMTs.
El-Osta et al. showed that transient exposure to hyperglycaemia causes persistent epigenetic changes in the promoter of nuclear factor κB in endothelial cells [10]. The DCCT showed that early exposure to hyperglycaemia prolongs negative effects, including diabetic complications, during subsequent periods of improved glycaemia [9]. Transient ischemia also decreased histone acetylation. An initial decrease in HAT activity occurred in the ischemic period, followed by the downregulation of HDAC5, partially contributing to histone reacetylation and BMP7 induction in the recovery period [35]. Histone methylation is a dynamic process. Lysine demethylase 1 specifically removes H3K4me marks [36] and is subsequently able to remove H3K9me [36]. These results showed that transient changes in conditions can result in epigenetic changes.
In conclusion, transient and persistent HG affects inflammatory gene expression and histone-modifying gene expression in RMCs. Considering the dynamic regulation of histone modifications and transient effects on histone modifications, cell-specific and disease stage-specific approaches targeting histone modifications should be a focus of future research.
Acknowledgements
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2010-013-E00009) and the Inje Research and Scholarship Foundation in 2010.
Conflict of Interest
All authors declare no conflict of interest.
References
- Thomas MC, Brownlee M, Susztak K, Sharma K, Jandeleit-Dahm KA, Zoungas S. Diabetic kidney disease. Nat Rev Dis Primers 2015; 30: 15018
- Navarro-González JF, Mora-Fernández C, Muros de Fuentes M, Garcia-Perez J. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol 2011; 7: 327-340
- Chow F, Ozols E, Nikolic-Paterson DJ, Atkins RC, Tesch GH: Macrophages in mouse type 2 diabetic nephropathy: correlation with diabetic state and progressive renal injury. Kidney Int 2004; 65: 116-128.
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 2002; 3: 224-229
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009; 10: 32-42.
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 3; 13: 343-357.
- Thomas MC. Epigenetic mechanisms in diabetic kidney disease. Curr Diab Rep 2016 ; 16: 31.
- Li X, Li C, Sun G. Histone acetylation and its modifiers in the pathogenesis of diabetic nephropathy. J Diabetes Res 2016; 2016: 4065382.
- Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, Raskin P, Zinman B. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med 2005; 353: 2643-2653.
- El-Osta A, Brasacchio D, Yao D. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008; 205: 2409-2417.
- Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 2010; 59: 2904-2915.
- Reddy MA, Tak Park J, Natarajan R. Epigenetic modifications in the pathogenesis of diabetic nephropathy. Semin Nephrol 2013; 33: 341-353.
- Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S, Tikoo K, Anders HJ. Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant 2010; 25: 1811-1817.
- Na SY, Kang BY, Chung SW, Han SJ, Ma X, Trinchieri G, Im SY, Lee JW, Kim TS. Retinoid inhibit interleukin-12 production in macrophages through physical association of retinoid X receptor and NF kappaB. J Biol Chem 1999; 274: 7674-7680.
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6: 838-849
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007; 25: 1-14.
- Giguere V. Retinoic acid receptors and cellular retinoid binding proteins; complex interplay in retinoid signaling. Endocr Rev 1994; 15: 61-79.
- Deb DK, Bao R, Li YC. Critical role of the cAMP-PKA pathway in hyperglycemia-induced epigenetic activation of fibrogenic program in the kidney. FASEB J 2017; 31: 2065-2075.
- Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, Natarajan R. Involvement of p300/CBP and epigenetic histone acetylation in TGF-β1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol 2013; 304: 601-613.
- Wang X, Liu J, Zhen J, Zhang C, Wan Q, Liu G, Wei X, Zhang Y, Wang Z, Han H, Xu H, Bao C, Song Z, Zhang X, Li N, Yi F. Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int 2014; 86: 712-725.
- Wei Q, Dong Z. HDAC4 blocks autophagy to trigger podocyte injury: non-epigenetic action in diabetic nephropathy. Kidney Int 2014; 86: 666-668.
- Gaikwad AB, Gupta J, Tikoo K. Epigenetic changes and alteration of Fbn1 and Col3A1 gene expression under hyperglycaemic and hyperinsulinaemic conditions. Biochem J 2010; 432: 333-341.
- Noh H, Oh EY, Seo JY, Yu MR, Kim YO, Ha H, Lee HB. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-1-induced renal injury. Am J Physiol Renal Physiol 2009; 297: 729-739.
- Advani A, Huang Q, Thai K, Advani SL, White KE, Kelly DJ, Yuen DA, Connelly KA, Marsden PA, Gilbert RE. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. Am J Pathol 2011; 178: 2205-2214.
- Gilbert RE, Huang Q, Thai K, Advani SL, Lee K, Yuen DA, Connelly KA, Advani A. Histone deacetylase inhibition attenuates diabetes-associated kidney growth: potential role for epigenetic modification of the epidermal growth factor receptor. Kidney Int 2011; 79: 1312-1321
- Ghosh AK, Yuan W, Mori Y, Varga J. Smad-dependent stimulation of type I collagen gene expression in human skin fibroblasts by TGF-beta involves functional cooperation with p300/CBP transcriptional coactivators. Oncogene 2000; 19: 3546-3555.
- Yoshikawa M, Hishikawa K, Marumo T, Fujita T. Inhibition of histone deacetylase activity suppresses epithelial-to-mesenchymal transition induced by TGF-beta1 in human renal epithelial cells. J Am SocNephrol. 2007; 18: 58-65.
- Cai M, Bompada P, Atac D, Laakso M, Groop L, De Marinis Y. Epigenetic regulation of glucose-stimulated osteopontin (OPN) expression in diabetic kidney. BiochemBiophys Res Commun 2016; 469: 108-113.
- Reddy MA, Sumanth P, Lanting L, Yuan H, Wang M, Mar D, Alpers CE, Bomsztyk K, Natarajan R. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int 2014; 85: 362-373.
- Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem 2008; 283: 26771-26781.
- Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 2010; 21: 2069-2080.
- Chen J, Guo Y, Zeng W, Huang L, Pang Q, Nie L, Mu J, Yuan F, Feng B. ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. Am J Physiol Renal Physiol 2014; 306: 916-925.
- Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl AcadSci USA 2008; 105: 9047-9052.
- Yuan H, Reddy MA, Deshpande S, Jia Y, Park JT, Lanting LL, Jin W, Kato M, Xu ZG, Das S, Natarajan R. Epigenetic histone modifications involved in profibrotic gene regulation by 12/15-lipoxygenase and its oxidized lipid products in diabetic nephropathy. Antioxid Redox Signal 2016; 24: 361-375.
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6: 838-849.
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007; 25: 1-14.