ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 15
HIV-1 induced differential expression of cell surface proteins in SupT1 cell lines
Parikipandla Sridevi1*#, Balakrishna SL2# and Bharti Gupta1
1Department of Biotechnology, Faculty of Science, Indira Gandhi National Tribal University, Amarkantak, Madhya Pradesh, India
2Department of Biotechnology and Bioinformatics, School of Life Sciences-Centre for Advanced Studies, University of Hyderabad, P. O. Central University, Hyderabad, Telangana, India
#These authors have equally contributed.
- *Corresponding Author:
- Parikipandla Sridevi
Department of Biotechnology
Faculty of Science
Indira Gandhi National Tribal University
Amarkantak, Madhya Pradesh, India
Accepted date: July 16, 2018
DOI: 10.4066/biomedicalresearch.29-18-723
Visit for more related articles at Biomedical ResearchObjective: HIV-1 infection to cluster of differentiation 4 (CD4)+ T cells alters many cellular pathways in host genome by protein-protein interactions.
Methods: To better understand these events of infection and change in host protein expression, we opted an approach of differential protein expression profiling of CD4+ T cell; SupT1 cells. We studied the change in expression of surface proteome after HIV-1 induction in SupT1 cell line, after 4 d post infection. The level of protein expression was checked by isolating the surface proteins from SupT1 cell culture by biotinylating and immunoprecipitation followed by gradient SDS-PAGE (2%-20%).
Results: A protein of 120 kDa found to express in the infected cell line when compared with the control. Protein extracted was further subjected to 2D gel electrophoresis, three proteins were found to be differentially expressing in HIV-1 infected cells.
Conclusion: Further analysis of these newly expressed proteins, not only helps in understanding the altered expression of surface proteins in CD4+ T cells after HIV infection but also in identification of potential molecules to be used in targeted drug delivery.
Keywords
HIV-1, CD4+ T cells, Cell surface markers, CD4, CCR4, CXCR5, Proteome analysis.
Introduction
HIV-1 enters into cell by the binding of the viral glycoproteins (gp120 and gp41) to the cellular receptor and co-receptor, leading to fusion of viral and cellular membranes. Cluster of differentiation 4 (CD4) is a type I transmembrane glycoprotein and functions in T cells as a receptor for HIV-1 binding [1]. HIV entry begins with the adhesion of virus to the host cell and ends with the fusion of the cell and viral membranes with delivery of the viral core into the cytoplasm [2,3]. Once viral core enters into cytoplasm, it gets disintegrated and releases Viral RNA which further processed to DNA by reverse transcription. The viral DNA in the form of pre-integration complex transported into the nucleus to become provirus which produces progeny virus by undergoing transcription and translation using host machinery; For successful viral production, host restriction factors get surpassed and the machinery which supports viral processes should be enhanced. This entire process is based on the modulation of the host proteins in response to the viral infection. Each step of viral lifecycle involved with signal transduction process which modulates complex mechanisms starting from the beginning, which are not yet fully explored. Upon the receptor CD4 [4] binding by the gp120 subunit [5,6] of viral Envelope, lot of rearrangements occurs on host cell surface. To the site of viral binding, co-receptors (CXCR4/CCR5) get transported which are required for viral fusion process. All these three glycoproteins of host cell surface are capable of transmitting signals results in change in proteome of the of host cell. Cell signalling modulations can happen in host by itself or by the influence of viral regulatory proteins released into host by virus or encoded in host genome which expires at later stages. A number of HIV-specific studies include analyses of the proteome of infected T cells [7], macrophages [8], intact HIV particles, purified HIV cores and HIV reverse transcription and pre-integration complexes [9]. Viral and host protein interaction has also been studied, including the viral proteins Gag [10], Nef [11], Tat [12], and Rev [13], as well as a comprehensive study of all HIV proteins.
Sup-T1 cells are of the CD4+ cell lines well adapted in laboratory conditions and the viral strain used in this study was 93IN101 which is a dual tropic virus and infection of Sup-T1 cells with this viral strain was well established in laboratory.
These cells express CXCR4 receptor and no traces of CCR5 on cell surface provide a clear infection rout which reduces ambiguity in the study and analysis.
Our goal of this study was the expression of cell surface proteins induced by HIV-1 infection in CD4+ cell lines using a sub-cellular focused proteomic approach. Sup-T1 cells were infected with HIV-1 virus and surface proteins were isolated using biotinylating process as described earlier [14]. Cell lysates were also separated into cytosolic, membrane bound, and nuclear fractions by differential lysis. 1D SDS?PAGE, followed by 2D-DIGE, were used. Intact protein fractionations used in this HIV biomarker study. These methods also have been used in several studies to discover biomarkers in body fluids of HIV?infected persons [15,16].
Our study is an effort to fill up the gaps in knowledge of early changes of host proteins after HIV infection. In this study, we described the results in the change of proteome of infected cells. By infecting SupT1 cell lines in vitro and assessing the differences in their surface protein expression over time, we aimed to cover the early changes caused by HIV in CD4+ T cells (SupT1 cells) proteome. Analysis of these newly expressed proteins helps in understanding the altered expression of surface proteins in CD4+ T cells after HIV infection and to identify potential molecules to be used in targeted drug delivery.
Materials and Methods
Cell and virus culture
All procedures involving infectious HIV-1 was carried out in biosafety level (BSL) 2 labs with BSL 3 practices. The SupT1 cells were propagated in RPMI (Gibco-BRL, Invitrogen, CA, USA) with 10% FBS (Gibco-BRL) at standard conditions of 5% CO2 and 37°C. HIV-1 93IN101 (subtype C) was cultured by infecting the SupT1 cells with cryopreserved viral stock in the presence of 8 μg/ml Polybrene (Sigma, St. Louis, MO, USA) overnight. The unbound virus was removed by washing the cells with 0.1 M phosphate buffered saline (PBS) three times and the cells were seeded back to complete medium. After 4 d of infection, the shedding virus was quantified in the culture supernatant with p24 ELISA kit (ABL Inc, Kensington, MD, USA), filtered using 0.45 μ membrane filter (Pall Corporation, USA) and used for infection experiments. Infection was given at 20 ng (100 MOI) viruses for every 2 × 106 cells in the presence of 8 μg/ml polybrene in serum free media.
HIV-1 p24 antigen assay
Cells were infected with HIV-1 for 2 h in the serum free media. The unbound virus was washed with 0.1M PBS, three times and the cells were seeded back in the complete medium. After 4 d of infection, the shedding virus was quantified in the free culture supernatants with p24 ELISA kit following manufacturer’s instructions.
Biotinylation of the cell surface proteins
All solutions employed were cooled to 4°C. Adherent monolayers of SupT1 cells in flask (2 × 107 cells cells/flask) were washed once with PBS pH 7.4. 15 mL of PBS containing 490 mM sulfo-NHS-SS-biotin were added to flask and cells were incubated at room temperature for 5 min. The biotinylation reaction was terminated by adding Tris-Cl to a final concentration of 4.9 mM. The cells were washed once and harvested in 10 mL of 490 mM oxidized glutathione solution in PBS by scraping.
Isolation of biotinylated proteins: The cells were pelleted and lysed for 30 min on ice with 1 ml of lysis buffer containing 2% w/v NP-40 substitute (Fluka, Buchs, Switzerland), 0.2% w/v SDS (Amersham Biosciences, Uppsala, Sweden), 100 mM oxidized glutathione, and protease inhibitors (Complete, EDTA-free from Roche Diagnostics, Basel, Switzerland). The cell lysate was cleared by centrifugation (16 100 × g, 10 min) followed by the purification of biotinylated proteins on streptavidin Sepharose high performance (Amersham Biosciences). 640 μl of streptavidin sepharose high performance were washed three times with wash buffer A (1% w/v NP-40 substitute and 0.1% w/v SDS in PBS) before adding the cleared cell lysates. The samples were incubated with streptavidin Sepharose high performance for 2 h on ice. Unbound proteins were removed by washing three times with wash buffer A, two times with wash buffer B (0.1% NP-40 w/v substitute and 0.5 M NaCl in PBS), and once with 50 mM Tris- HCl, pH 7.5.
Elution of captured proteins: Elution of captured proteins from streptavidin sepharose was done by incubating with 0.4 mL of 5% 2-mercaptoethanol (Sigma-Aldrich) in PBS for 30 min at 30°C. The elution step was repeated three times. The collected elutes were pooled and the eluted proteins were precipitated by adding 200 mL TCA (Fluka) to a final concentration of 10% and incubating elutes for 30 min on ice. The precipitate was pelleted by centrifugation (10,000 × g, 5 min), washed with 1 ml of ethanol-ether (1:1) and air dried.
Digestion of the eluted proteins: Elution of digested proteins was done by dissolving the TCA precipitate in 200 ml of 50 mM ammonium hydrogen carbonate solution and heated to 95ºC for 10 min. The solution was cooled on ice and trypsin (Promega, Madison, WI, USA) was added to a concentration of 60 ng/mL; the digestion was carried out for 15 h at 37°C followed by drying the samples in a SpeedVac concentrator and storing them at -20°C until use for SDS PAGE electrophoresis.
Gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
100 μg of each protein sample was used to perform SDSPAGE (10%) and sample was prepared by mixing protein with the sample buffer containing 0.125 M Tris-Cl (pH 6.8), 4% SDS, 20% glycerol, 10% 2-mercaptoethanol and 0.002% bromophenol blue followed by incubation at 100°C for 5 min. Tris-glycine buffer (25 mM Tris and 192 mM glycine (pH 8.3)) with 0.1% SDS was used as the electrode buffer and electrophoresis was carried out at 100 V. This was carried according to the standard silver staining protocol. The gel was incubated in fixative (50% methanol, 12% acetic acid and 50 μl of 37% formaldehyde/100 ml) for 1 h followed with 3 washes in 50% ethanol. Subsequently the gel was pre-treated with sodium thiosulphate (20 mg/100 ml) for 1 min and rinsed thrice (20 s each) with distilled water. The gel was impregnated with silver nitrate (0.2% containing 187 μl of 37% formaldehyde) with gentle agitation for 30 min. It was rinsed with distilled water and developed with 6% sodium carbonate (w/v) and 50 μl of 37% formaldehyde (v/v). Finally, the stained gel was thoroughly rinsed with distilled water and stored in 50% methanol.
2D gel electrophoresis
Total protein was prepared by lysing infected and uninfected cells in protein extraction buffer (10 mM Tris-Cl (pH 7.4), 0.1% Triton X-100, 1 mM PMSF, 1 mM EDTA and 1 mM DTT) for 10 min on ice with protease inhibitor cocktail (GE Healthcare, UK). The lysates were centrifuged at 12000 × g for 15 min at 4°C; supernatants collected were quantified by Bradford method. The supernatants collected were subjected to clean-up using the 2-DClean-up Kit (GE Healthcare Life Sciences, USA) as per the manufacturer’s instructions.
The clean-up process was followed by dissolution of the separated protein precipitate in rehydration buffer constituting 7 M urea, 2 M thiourea, 4% CHAPS, 2% ampholytes of pH 3-10, 70 mM DTT and 3.5% bromophenol blue. IPG strips (Immobiline Drystrip, 3-10 pH linear 18 cm, GE Healthcare) were rehydrated in 350 L of the protein solution for 20 h under low viscosity mineral oil (Merck Genei, Bangaluru, India). The rehydrated IPG strips containing 300 g protein were then subjected to Isoelectric Focusing in Ettan IP Gphor3 gel apparatus (GE Healthcare) at 20°C for 70,000 Vh-50 V-1 h, 500 V-5 h, 500 V-5 h, 10000 V-8 h, 500 V-10 h (a total of 70,000 Vh). After IEF, the strips were equilibrated in equilibration buffer (75 mM Tris-HCl pH 8.8, 30% glycerol, 2% SDS and a trace of bromophenol blue) containing 10 mg/ml of DTT followed by 20 min in equilibration buffer containing 25 mg/ml of iodoacetamide (an alkylating agent to reduce cysteine residues preventing disulfide bond formation). SDS-PAGE was carried out and the gels were then silver stained according to the standard silver staining protocol. The gel was incubated in fixative (50% methanol, 12% acetic acid and 50 μl of 37% formaldehyde/100 ml) for 1 h followed with 3 washes in 50 % ethanol. Subsequently the gel was pre-treated with sodium thiosulphate (20 mg/100 ml) for 1 min and rinsed thrice (20 s each) with distilled water. The gel was impregnated with silver nitrate (0.2% containing 187 μl of 37% formaldehyde) with gentle agitation for 30 min. It was rinsed with distilled water and developed with 6% sodium carbonate (w/v) and 50 μl of 37% formaldehyde (v/v). Finally, the stained gel was thoroughly rinsed with distilled water and stored in 50% methanol.
Statistical analysis
All statistical analyses were done using the SPSS 10.0 software package (SPSS Inc., Chicago, IL, USA). Differences between groups were compared using Anova and T tests. All tests were two-tailed and were considered significant at P<0.1.
Results
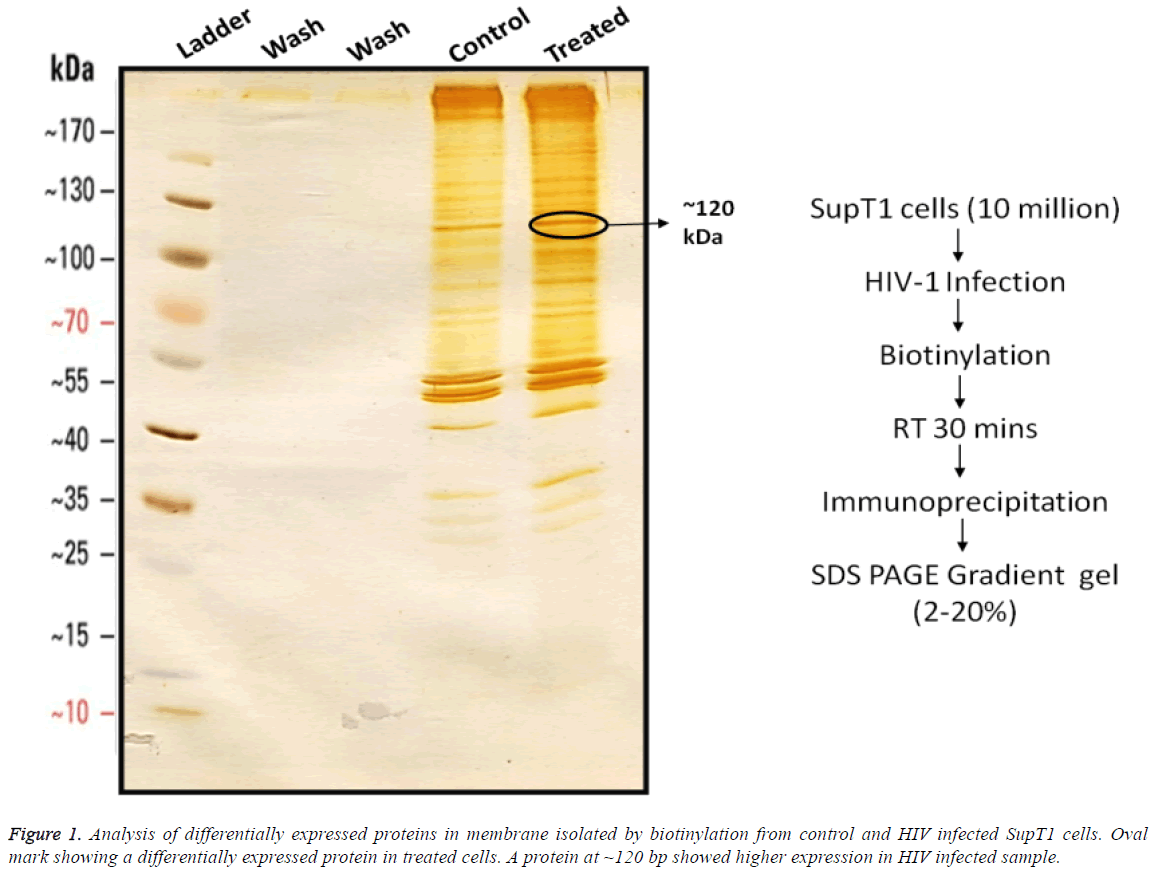
Differentially expressed proteins on SupT1 cells were isolated by biotinylation followed by gradient SDS-PAGE (2%-20%) from control and HIV infected cells (Figure 1). Cell surface proteins from Infected and control cells were biotinylated and immuno-precipitated using streptavidin conjugated beads as mentioned in materials and methods. Results show that there was no detachable amount of protein present in wash through from control and infected samples. It indirectly represents the efficiency of biotin-avidin based immunoprecipitation system. Immunoprecepitate from both samples has produced clear sharp protein bands represent a good resolving of proteins on gel. In comparison, the immunoprecipitate from infected cells has produces more protein bands than control. Even though several proteins seemed to be expressed differentially, a protein of 120 kDa in SupT1 cells showed higher expression.
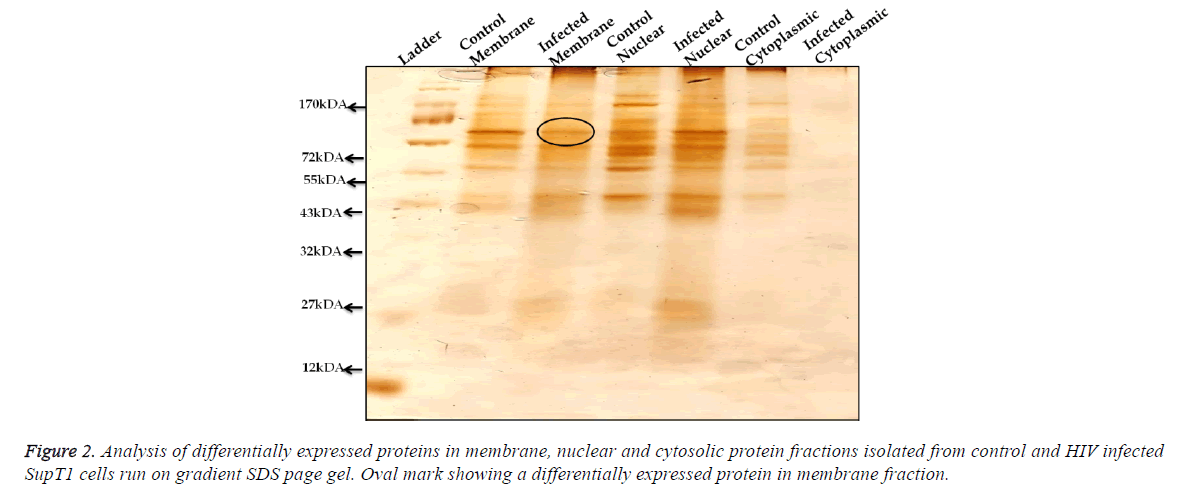
Analysis of differentially expressed proteins in membrane, nuclear and cytosolic protein; from control and HIV-1 infected SupT1 cells fractions of membrane proteins, cytosolic and nuclear proteins were isolated and these fractions were run on gradient SDS-PAGE gel (5%-20%) (Figure 2) and silver stained. Results show that HIV-1 infection leads to the differential expression of proteins and was observed not only in membrane fraction but also in nuclear and cytosolic fractions of infected cell lysates.
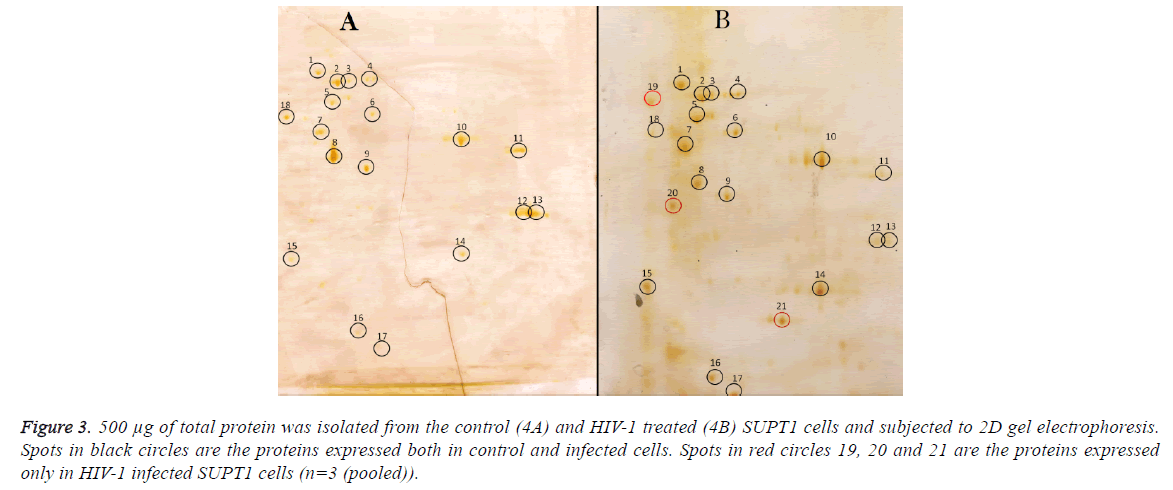
Comparison of differentially expressed proteins in 2D gel electrophoresis; 250 μg proteins from control cells were run on 2D gel electrophoresis and 18 spots were observed on the gel (Figure 3), same results were seen with 500 μg protein from control sample. 500 μg of total protein was isolated from the control (3A) and HIV-1 treated (3B) SupT1 cells and subjected to 2D gel electrophoresis. Out of 21 spots, 15 proteins differentially expressed, and remaining has no effect of HIV-1 infection.
Proteins corresponding to spot no 5, 8, 11, 12, 13 and 18 expression levels were decreased and expression of proteins with spot no 6, 10, 14, 15, 16 and 17 were up regulated and three protein spots appeared newly (19, 20 and 21) and there was no detectable difference observed in remaining proteins upon HIV-1infection.
Discussion
Several proteomics approaches like 1D gel electrophoresis, 2D gel electrophoresis, MALDI-TOFF, LC-MS/MS [17] etc. have implicated to study the proteomics changes after HIV infections to CD+ T cells in vitro. Some have studied the changes in the proteome of human CD4+ T cells associated with HIV-1 infection both in vitro and in vivo, and found changes in the overall proteome of CD4 T cells infected with HIV-1 in vitro after 24 h and also studied the involvement of some important functional groups within the cell [18]. In addition to qualitative analysis of cell surface protein some also measured the quantity and dynamics of certain HIV-1 proteins within the host proteome [19]. In our work we studied the change in cell surface protein expression after 4 d post infection.
In this study, we utilized a high-throughput quantitative proteomic analysis to identify changes in host cell protein upon infection of SupT 1 cell line with HIV-1. To find out the differential expression of the proteins whose expression can be influenced by HIV-1 infection were tested at membrane, nuclear and cytoplasmic levels by fractionation and obtained data revealed that all these three fractions contain differentially expressed proteins (Figure 2) and HIV-1 infection has impact on several proteins involved in diverged cellar processes. Similar differential expression results at early hours of post infection were reported [20,21] with an observation of early hours of infection resulted in differential expression of maximum proteins and when time increases the proteins with differential expression decreases [21].
In other studies, among the most significantly affected cellular functions were protein early up-regulation, followed by downregulation of some cell surface proteins involved in cellular proliferation, T cell immune function maintenance and [18] DNA repair and recombination processes. These changes, some of which may have occurred very early after infection of HIV-1 to CD4+ T cells, these studies may suggest some novel strategies by which the HIV-1 virus manipulates host cell machinery to its own advantage and changes the protein expression of host cells [22]. Further experimental evaluation of differential expression specific to membrane protein revealed several proteins were differentially expressed and a prominent overexpression of a protein with molecular weight approx. 120 kDa was observed in gradient SDS-PAGE (Figure 1). Further analysis of this differentially expressed proteom in 2D gel electrophoresis by comparing control and infection showed 21 different spots were observed in infection condition, out of them 3 were newly appeared (Figure 3B). These three new proteins may be the result of HIV-1 infection and indicating the host gene expression under the influence of infection. Along with these 12 other proteins have shown differential expression. Out of them expression of 6 proteins were enhanced and 6 proteins were suppressed similar down regulated results were reported earlier [23] and viral accessory proteins Vpu or Nef involvement in these effects by disrupting intra cellular transport mechanisms in host cells [24] the accessory proteins Nef, Vif, Vpu, and Vpr interact with different proteins and pathways of the cells of immune system to bypass host cell mediated antivirus resistance [25].
Figure 3: 500 μg of total protein was isolated from the control (4A) and HIV-1 treated (4B) SUPT1 cells and subjected to 2D gel electrophoresis. Spots in black circles are the proteins expressed both in control and infected cells. Spots in red circles 19, 20 and 21 are the proteins expressed only in HIV-1 infected SUPT1 cells (n=3 (pooled)).
Host factors not only exert a negative influence but also exert positive influence [9] on the viral infection like LEDGF (lens epithelium-derived growth factor) (HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells., an essential role for LEDGF/p75 in HIV integration), CDK9/Cyclin T1 (A novel CDK9-associated Ctype cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA), TSG101 (Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding) and topoisomerase II [26] involved in various viral process.
Significant appearance of three proteins expression in HIV infected cell when compared to control may have potential to become biomarker for HIV-1 infection in Sup-T1 cells. Further characterization of these three proteins followed by their identification by MALDI-Toff was required.
To summarize, analysis of differential expression of proteins by 2D gel electrophoresis showed marked difference in their protein pattern in HIV-1 infection. Three proteins showed significant expression in HIV-1 infected cells when compared to control which can be further analyzed by MALDI-Toff. Identification and characterization of the newly expressed proteins may reveal the possibility of using them in targeted drug delivery.
Acknowledgement
PSD gratefully acknowledges UGC Dr. Kothari Post-Doctoral Research Fellowship from University Grants Commission, Govt. of India and SLBK acknowledges DST Research Associate fellowship. We thank Prof. Anand K. Kondapi, University of Hyderabad for extending facilities for these studies and funds from the DST are acknowledged.
References
- Saphire AC, Bobardt MD, Zhang Z, David G, Gallay PA. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J Virol 2001; 75: 9187-9200.
- Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA. HIV-1 envelope protein binds to and signals through integrin α4β7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 2008; 9: 301-309.
- Cecilia D, Kulkarni SS, Tripathy SP, Gangakhedkar RR, Paranjape RS, Gadkari DA. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 2000; 271: 253-258.
- Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 1986; 47: 333-348.
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS Res Hum Retroviruses 2005; 21: 171-189.
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 2002; 420: 678-682.
- Ringrose JH, Jeeninga RE, Berkhout B, Speijer D. Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J Virol 2008; 82: 4320-4330.
- Haverland NA, Fox HS, Ciborowski P. Quantitative proteomics by SWATH-MS reveals altered expression of nucleic acid binding and regulatory proteins in HIV-1-infected macrophages. J Proteome Res 2014; 13: 2109-2119.
- Lokeswara SB, Anand KK. Role of host proteins in HIV-1 early replication, Advances Mol Retrovirol 2016.
- Le Sage V, Cinti A, Valiente-Echeverria F, Mouland AJ. Proteomic analysis ofHIV-1 Gag interacting partners using proximity-dependent biotinylation. Virol J 2015; 12: 138.
- Mukerji J, Olivieri KC, Misra V, Agopian KA, Gabuzda D. Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology 2012; 9: 33.
- Coiras M, Camafeita E, Ureña T, López JA, Caballero F, Fernández B, López- Huertas MR, Pérez-Olmeda M, Alcamí J. Modifications in the human T cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics 2006; 6: 63-73.
- Naji S, Ambrus G, Cimermancic P, Reyes JR, Johnson JR, Filbrandt R, Huber MD, Vesely P, Krogan NJ, Yates JR. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol Cell Proteomics 2012.
- Rozek W, Ricardo-Dukelow M, Holloway S, Gendelman HE. Cerebrospinal fluid proteomic profiling of HIV-1-infected patients with cognitive impairment. J Proteome Res 2007; 6: 4189-4199.
- Rozek W, Horning J, Anderson J, Ciborowski P. Sera proteomic biomarker profiling in HIV?1 infected subjects with cognitive impairment. Proteomics Clin Appl 2008; 2: 1498-1507.
- Kim SS, Kim MH, Shin BK, Na HJ. Different isoforms of apolipoprotein AI present heterologous post?translational expression in HIV infected patients. J Proteome Res 2007; 6: 180-184.
- Loyda MM, Krystal C, Linda R, Eillen R-F, Dianedis T-N. Proteomic analysis of HIV-infected macrophages. J Neuroimmune Pharmacol 2011; 6: 89-106.
- Arti T, Pavel S, David E, Jeffrey M, Alejandro W, Marcus J, Stewart T, Sean C, Tahmina A. Quantitative proteomic analysis of HIV-1 infected CD4þ T cells reveals an early host response in important biological pathways: Protein synthesis, cell proliferation, and T-cell activation. Virology 2012; 429: 37-46.
- Gonza´ lez-Navajas JM, Fine S, Law J, Datta SK, Nguyen KP, Yu M, Corr M, Katakura K, Eckman L, Lee J, Raz E. TLR4 signaling in effector CD4_ T cells regulates TCR activation and experimental colitis in mice. J Clin Invest 2010; 120: 570-558.
- Johannes N, Valentina V, Karin JM, Victoria S, Rainer W, Patrick P, Ruedi A, Huldrych G, Ben CC. In vivo and in vitro proteome analysis of HIV-1 infected human CD4+ T cells. Mol Cell Proteomics 2017.
- Arti TN , Pavel S, David EP, Jeffrey MW, AlejandroW-Y, Marcus JK, Stewart TC , Sean CP , Tahmina AJ, Alexei LK , Robert EP , Michael GK . Quantitative proteomic analysis of HIV-1 infected CD4+ T cells reveals an early host response in important biological pathways: Protein synthesis, cell proliferation, and T-cell activation. Virology 2012; 429: 37-46.
- Krishna KD, Santanu B, Mohan H, Owen W, Viswanath R, Sherwin L, Indira H. Analysis of host gene expression profile in HIV-1 and HIV-2 infected T cells. PLoS One 2016; 11: 1.
- Nicholas JM, Jonathan S, Kim W, Clary BC, Stuart JDN, Paul JL. Cell surface proteomic map of HIV infection reveals antagonism of amino acid metabolism by Vpu and Nef. Cell Host Microbe 2015; 18: 409-423.
- Claudia H, Birthe M, Joëlle VF, Miguel L-M, Bettina S, François MP, Oliver TK, Oliver TF, HIV-1 Nef and Vpu are functionally redundant broad-spectrum modulators of cell surface receptors, including tetraspanins. J Virol 2014; 88: 14241-14257.
- Michael HM, Michael E. HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host Microbe 2008; 3: 388-398.
- Lokeswara SB, Nathamu S, Anand KK. Involvement of human topoisomerase II isoforms in HIV-1 reverse transcription. Archive Biochem Biophys 2013; 532: 91-102.