ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 3
Identification and phylogenetic analysis of the sheep pox virus Shanxi isolate
Shao-peng Gu1*, Xin-tao Shi2, Xing-guo Liu3, Zhong-bing Wang4, Ming-xue Zheng1, Yun Li2, Nai-rui Huo1, Fang Tang1, Rui Bai1, Zong-yong Shi5 and Jian-qin Yuan5
1College of Animal Science and Veterinary of Shanxi Agricultural University, Shanxi, Taigu County, PR China
2Center for Animal Disease Control and Prevention of TaiYuan, PR China
3Center for Animal Disease Control and Prevention of China, PR China
4Center for Animal Disease Control and Prevention of the Shanxi Province, PR China
5Genetically Modified Testing Center of Ministry of Agriculture and College of Life Science of Shanxi Agricultural University, Taigu, PR China
- *Corresponding Author:
- Shao-peng Gu
College of Animal Science and Veterinary of Shanxi
Agricultural University
No.1, Mingxian Road
Taigu County, Shanxi Province 030801, PR China
Accepted on November 8, 2017
DOI: 10.4066/biomedicalresearch.29-17-431
Visit for more related articles at Biomedical ResearchA strain of virus was successfully isolated from a Demei sheep with typical symptoms of sheep pox virus (SPPV) in Shanxi province in China (SPPV-Shanxi) and identified using hematoxylin and eosin staining, electron microscopy, neutralization testing, and PCR. Identification results proved a SPPV-Shanxi infection in the sheep. The full-length P32 gene (an immunogenic envelope protein of Capripoxvirus) and the SPV64 gene (the L5R allele in SPPV) were cloned and sequenced. Phylogenetic trees were constructed based on the nucleotide and amino acid sequences of the two genes. Based on the phylogenetic tree, the SPPV-Shanxi isolate clustered together with AV40 (a Chinese vaccine strain) was closer to FJ748487 (SPPV India isolate) and AY077834 (SPPV Kazakhstan isolate). The amino acid sequence analysis revealed several unique amino acid substitutions between the SPPV-Shanxi isolate and other isolates. This is the first report that an outbreak of SPPV occurred in Shanxi province in China. There are no reports on the molecular characterization of SPPV-Shanxi isolates based on P32 and SPV64 genes. Our findings may provide the information on the genotype of the etiologic prevalence responsible for the SPPV outbreak in Shanxi province.
Keywords
Sheep poxvirus, Shanxi isolate, AV40, P32 gene, SPV64 gene.
Introduction
Sheep pox virus (SPPV) belongs to the Capripoxvirus genus of the Poxviridae family. Goat pox virus (GTPV) and Lumpy Skin Disease Virus (LSDV) of cattle [1] also are members of the Capripoxvirus genus. SPPV can cause significant economic losses if not properly controlled.
SPPV causes systemic diseases characterized by the fever, skin nodules, respiratory and gastrointestinal tract lesions, and lymph node enlargement. SPPV is a direct cause of high mortality [2], which approaches 100% in native animals [3] and sheep pox is categorized as a reportable disease by the World Organization for Animal Health (OIE).
Geographic regions affected by sheep pox and goat pox have been mainly limited to Africa and middle Asia in the last 50 years [4-7], but extended to Bangladesh in 1984 [8] and India [9,10], and more recently to Yemen, Vietnam [11] and Mongolia [12]. Repeated incursions have been reported in Greece and in southern Europe [1].
Capripoxviruses are double-stranded DNA viruses with genomes approximately 150 kb in size [13]. P32 is one of the structural proteins shared by all Capripoxviruses. P32 contains major antigenic determinants [14], which are important for the pathogenicity, diagnosis, prevention, and control of capripoxvirus infections. The fourth membrane protein, which exists in all poxviruses and is encoded by the L5R gene, is necessary for cell entry, cell-cell fusion, plaque formation, and infectious virion production [15]. The L5R allele in SPPV is the gene for a putative membrane protein (SPV64) [16], which has a function similar to the protein encoded by L5R and might be associated with viral infection and virulence.
AV40, a strain of SPPV, has been used for vaccine production in China since 1959 [17]. We reported previously that the morbidity and mortality reached 61% (183/300) and 15% (45/300), respectively, when a SPPV infection broke out in a herd of three hundred 1 y old non-vaccinated Demei sheep in Shanxi in 2010, which was identified as SPPV-Shanxi. The full-length P32 genes and partial SPV64 genes were amplified by PCR from SPPV-Shanxi and AV40, then phylogenetic analysis was performed among AV40, SPPV-Shanxi, and other 20 isolates. The findings may provide further information about the genotype of the SPPV prevalence in Shanxi province and the molecular characteristics of various SPPV strains.
Materials and Methods
Isolation of lamb testis cell
Nodular and papular lesions from infected sheep were washed with 1.5 ml of Phosphate Buffered Saline (PBS), then homogenized and directly inoculated to Lamb Testis (LT) cell preparations. Primary LT cells were isolated and cultured using routine cell culture techniques [18]. We harvested the virus by centrifugation, followed by repeated freezing and thawing of the culture. The virus was named SPPV-Shanxi and stored at -20°C as the antigen for all subsequent experiments.
Assessment of infection efficiency using H and E staining
LT cells were cultured in 24-well tissue culture plates with a 5 mm2 glass coverslip in each well, and infected with 200 μl of SPPV-Shanxi. When Cytopathic Effect (CPE) was observed, the cells were stained with H & E, then detected under a light microscope for cytoplasmic inclusions. Non-inoculated LT cells were used as negative controls.
Electron microscope studies
SPPV-Shanxi were stained using phosphotungstic acid, then applied to a carbon-coated formvar film on a 400-mesh copper grid. The grid was examined under a GEOL 1400 transmission electron microscope to investigate the surface structure of the viruses.
Neutralization testing
LT cells were grown on a 6-well tissue culture plate. The sheeppox antiserum (supplied by the China Institute of Veterinary Control) was diluted 1:5 in DMEM. Two hundred microliters of the virus were mixed with sheep pox antiserum in an equal volume (1:1), and 200 μl of standard sera, 200 μl of the SPPV-Shanxi, and 200 μl of DMEM were added separately to 4 wells. The plate was incubated at 37°C for 1 h, and observed under a microscope.
DNA extraction
The viral DNA was extracted using the E.Z.N.A. Mag-Bind Viral DNA/RNA Kit (Omega Bio-Tek, Inc.,) and eluted in 50 μl of DNA elution buffer according to the manufacturer’s instructions, then stored at -20°C until use.
PCR
Samples prepared as described above were used as templates. The primers were synthesized by AuGCT Biotechnology Synthesis Lab. The primers for SPPV-Shanxi P32 were as follows: forward, 5’- CGGGATCCACCATGGCAGATATCCCATTATA-3’; and reverse, 5’- GGAATTCCTAAATTATATACGTAAATAACATAC-3’). The forward and reverse primers for SPPV-Shanxi SPV64 were 5’- AACGAAAAGTTATCAGTC-3’ and 3’- GTTAATAGATACTTGCTG- 5’, respectively. The program for P32 and SPV64 amplification was as follows: 95°C denaturation for 10 min; 35 cycles at 95°C for 45 s, 56°C for 60 s (for SPV64, this step was at 49°C for 60 s), and 72°C for 80 s, then 72°C for 10 min. The DNA extracted from uninfected cells and supernatants were used as negative controls. Amplicons were visualized by electrophoresis in 1% agarose gels.
Cloning and DNA sequencing
The amplicons of the P32 and SPV64 genes were cloned into the pMD-18T vector (Takara, Dalian, China) and transformed into E. coli DH5α. The selected positive clones for sequencing were sent to TaKaRa Biotechnology (Dalian) Co., Ltd. The complete sequences of these two genes were submitted to the NCBI GenBank database and assigned accession numbers HM770955 and HM802211 (both are referred to as the Shanxi isolate), and the accession numbers for the AV40 strain were HQ607368 and HQ620556, respectively.
Phylogenetic analysis
Sequence comparison of SPPV-Shanxi to the other available Capripoxvirus sequences in the Genbank database was performed using the online BLAST program. Sequence identities of nucleotides, as well as those of amino acids, were analyzed using ClustalW2 Online. The deduced amino acid sequence was assembled into a multiple sequence alignment. A phylogenetic tree derived from nucleotide sequences was constructed for the Capripoxvirus using the maximum likelihood method of MEGA (version 5.0.1).
Results
Cytopathic effect
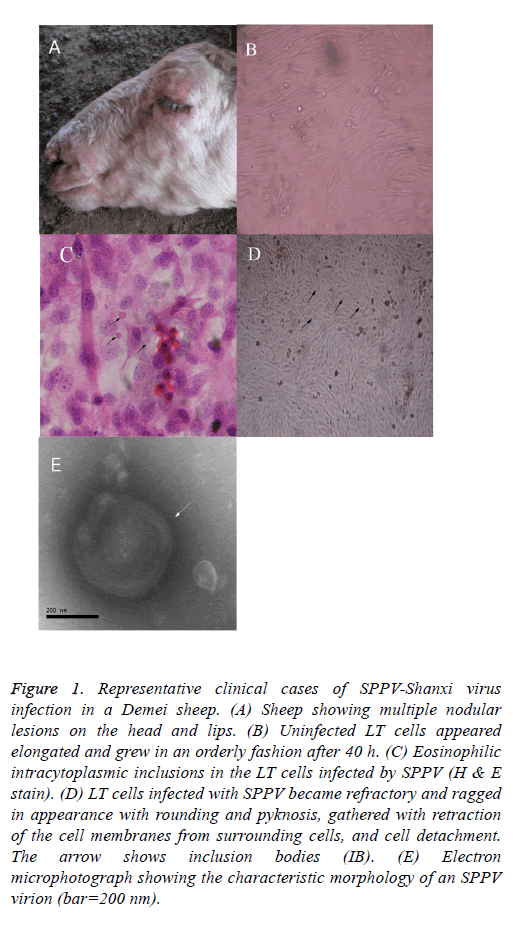
The clinical symptoms and the observed macroscopic lesions of the infected sheep were consistent with that of SPPV infection (Figure 1A). Infected LT cells appeared more elongated, grew in a more orderly style after 40 h of SPPV infection, and exhibited considerable morphologic heterogeneity (i.e., larger in size and eosinophilic intracytoplasmic inclusions in the infected LT cells; Figure 1B). Some LT cells showed symptoms of stress, featured by vacuolation and swelling. After experimental viral infection, the CPE was observed the 5th d after inoculation. Individual LT cells were refractory and had a ragged appearance, sphericity, and pyknosis, retraction of the cell membranes from surrounding cells, and cell detachment (Figure 1C (the arrow shows that the individual LT cells became spherical and larger in size, and inclusion bodies are noted)). On d 8, the CPE can be observed in most cells. The monolayer was destroyed as the infected cells became rounded. Eventually, 80% cells detached from the surface after 10 d. Most of the cells became disconnected from other cells and the culture fluid became turbid.
Figure 1: Representative clinical cases of SPPV-Shanxi virus infection in a Demei sheep. (A) Sheep showing multiple nodular lesions on the head and lips. (B) Uninfected LT cells appeared elongated and grew in an orderly fashion after 40 h. (C) Eosinophilic intracytoplasmic inclusions in the LT cells infected by SPPV (H & E stain). (D) LT cells infected with SPPV became refractory and ragged in appearance with rounding and pyknosis, gathered with retraction of the cell membranes from surrounding cells, and cell detachment. The arrow shows inclusion bodies (IB). (E) Electron microphotograph showing the characteristic morphology of an SPPV virion (bar=200 nm).
Virus identification by light and electron microscopy
After histological H&E staining, many eosinophilic intracytoplasmic inclusions were observed in the SPPVinfected LT cells (Figure 1C). Negatively-stained SPPV observed under electron microscopy revealed the presence of brick-shaped Orthopoxvirus particles 220-280 nm in length and 180-190 nm in width (Figure 1E). The virus particles had an oval- or rectangular-shaped particle containing a central core which ruled out the involvement of Parapoxvirus infection because parapoxviruses have superficial tubular structures [19].
Results of the neutralization test
Based on NT, after 5 d of incubation, with the exception of the wells inoculated with 200 μl of SPPV-Shanxi, the other three wells had negative results. This indicated the presence of SPPV antigen in the samples.
PCR results
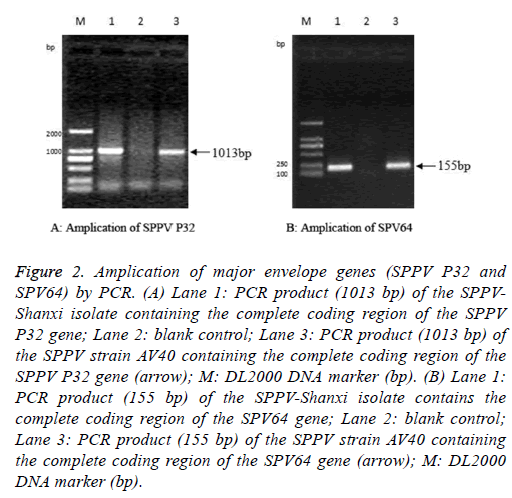
The full-length SPPV-Shanxi P32 gene, with an expected size of 1013 bp (Figure 2A), was amplified from the Shanxi isolate, the AV40 strain, and the CPE-positive LT cells. PCR also yielded an expected 155 bp amplification of the partial sequence of the SPV64 gene of the SPPV-Shanxi and AV40 strains (Figure 2B).
Figure 2: Amplication of major envelope genes (SPPV P32 and SPV64) by PCR. (A) Lane 1: PCR product (1013 bp) of the SPPVShanxi isolate containing the complete coding region of the SPPV P32 gene; Lane 2: blank control; Lane 3: PCR product (1013 bp) of the SPPV strain AV40 containing the complete coding region of the SPPV P32 gene (arrow); M: DL2000 DNA marker (bp). (B) Lane 1: PCR product (155 bp) of the SPPV-Shanxi isolate contains the complete coding region of the SPV64 gene; Lane 2: blank control; Lane 3: PCR product (155 bp) of the SPPV strain AV40 containing the complete coding region of the SPV64 gene (arrow); M: DL2000 DNA marker (bp).
Results of phylogenetic studies
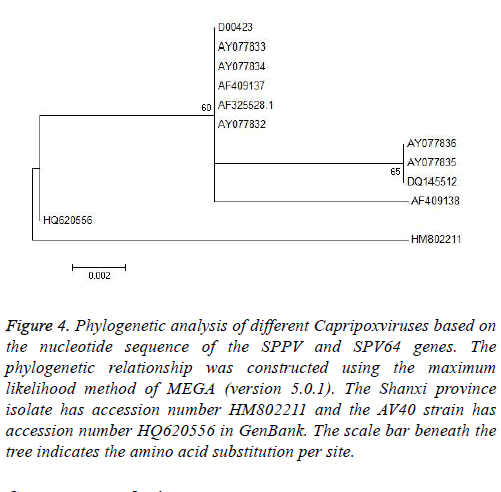
Phylogenetic analysis was performed using MEGA (version 5.0.1) and the maximum likelihood method. To determine the phylogenetic relationship between the full-length P32 and SPV64 genes, we compared DNA sequences obtained by PCR amplification with corresponding sequences, including SPPV, GTPV, and LSDV in the GenBank (Tables 1 and 2). The results of phylogenetic studies of the P32 and SPV64 gene showed that the SPPV-Shanxi and AV40 strains belonged to the same branch and SPPV-Shanxi was close to FJ748487 (SPPV, India, Figure 3) and AY077834 (SPPV, Kazakhstan; Figures 4). Additionally, the Shanxi isolate shared 97%~99% and 99%~100% identity with other SPPVs, 96%~98% positives and 97%~98% identity with GTPVs, and 96%~98% positives and 97%~98% identity with LSDVs. We conclude that the outbreak was caused by a SPPV.
| Gene accession number | Length (bp) | Species of Capripoxvirus | Country of isolation |
|---|---|---|---|
| HM770955 | 972 | SPPV | China |
| HQ607368 | 972 | SPPV | China |
| FJ748487.1 | 972 | SPPV | India |
| AY077834.1 | 972 | SPPV | Kazakhstan |
| AY368684.1 | 972 | SPPV | India |
| DQ153225.1 | 972 | SPPV | India |
| EU835938.1 | 972 | SPPV | India |
| DQ153223.1 | 928 | SPPV | India |
| DQ153219.1 | 918 | SPPV | Not available |
| AY077836.1 | 972 | GTPV | Kazakhstan |
| FJ748488.1 | 969 | GTPV | India |
| EU625263.1 | 969 | GTPV | Viet Nam |
| EF522177.1 | 969 | GTPV | China |
| EF514890.1 | 969 | GTPV | China |
| AY159333.1 | 969 | GTPV | India |
| EU625262.1 | 969 | GTPV | Yemen |
| EF514889.1 | 969 | GTPV | China |
| AF124516.1 | 969 | LSDV | Not available |
| AF325528.1 | 972 | LSDV | Not available |
| AF409137.1 | 972 | LSDV | South Africa |
| AF409138.1 | 972 | LSDV | South Africa |
Table 1. SPPV P32 genes of Capripoxviruses used in phylogenetic analysis.
| Gene accession number | Length (bp) | Species of Capripoxvirus | Country of isolation |
|---|---|---|---|
| HM802211 | 155 | SPPV | China |
| HQ620556 | 155 | SPPV | China |
| AF325528 | 155 | LSDV | Kenya |
| AF409137 | 155 | LSDV | Kenya |
| AY077834 | 155 | SPPV | Kazakhstan |
| AY077833 | 155 | SPPV | Kazakhstan |
| AY077832 | 155 | SPPV | Turkey |
| AY077836 | 155 | GTPV | Not available |
| AY077835 | 155 | GTPV | Not available |
| DQ145512 | 155 | GTPV | China |
| AF409138 | 155 | LSDV | South Africa |
Table 2. SPV64 genes of Capripoxviruses used in phylogenetic analysis.
Figure 3: Phylogenetic analysis of different Capripoxviruses based on the nucleotide sequence of SPPV P32 gene. The phylogenetic relationship was constructed using the maximum likelihood method of MEGA (version 5.0.1) The Shanxi province isolate has accession number HM770955 and the AV40 strain has accession number HQ607368 in GenBank. The scale bar beneath the tree indicates the amino acid substitution per site.
Figure 4: Phylogenetic analysis of different Capripoxviruses based on the nucleotide sequence of the SPPV and SPV64 genes. The phylogenetic relationship was constructed using the maximum likelihood method of MEGA (version 5.0.1). The Shanxi province isolate has accession number HM802211 and the AV40 strain has accession number HQ620556 in GenBank. The scale bar beneath the tree indicates the amino acid substitution per site.
Sequence analysis
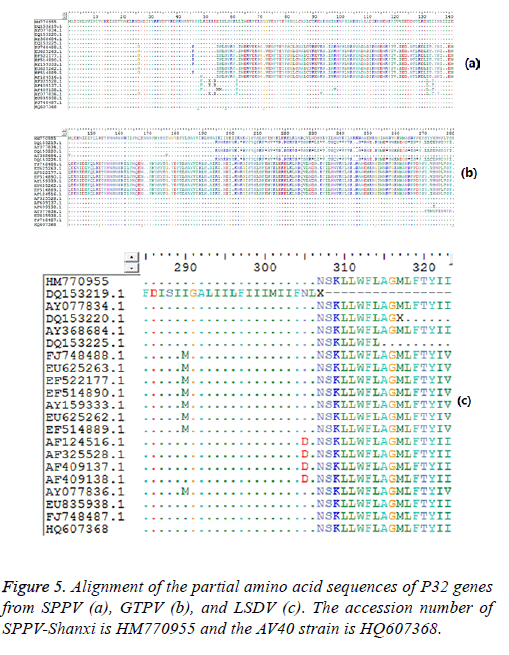
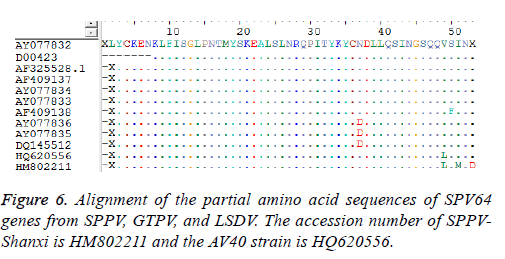
Sequence analysis results also demonstrated that the P32 and SPV64 genes between the SPPV-Shanxi isolate and the AV40 strain shared 98% and 99% identity, respectively, which indicated that the relationship was close. The length of the SPPV P32 gene (972 bp) was identical with the P32 gene of some SPPV isolates and the P32 gene of all cattle poxvirus isolates, but was 3 bp longer than most P32 genes of GTPV isolates Analyses based on the deduced amino acid sequences revealed 10 unique substitutions (D26, N46, D54, L62, A93, S132, I134, H136, I290, and I323) in the Shanxi isolate, which were sequentially G26, K46, none, F62, V93, L132, T134, Y136 (Figure 5a) M290 (Figure 5b), and V323 (Figure 5c) in other isolates, respectively. In the deduced amino acid sequence of the SPPV P32 gene of SPPV-Shanxi, 7 unique substitutions (F48, L58, G72, R153, E168, N253, and F281) were found, while in AV40 the unique substitutions were L48, I58, A72, D153, D168, Y253, and L281. There was only one amino acid difference between T145 in the SPPV-Shanxi and C145 in the AV40 strain. In addition, the deduced amino acid sequences of the P32 and SPV64 genes between the AV40 strain and other isolates showed few differences. In the case of the P32 gene, the differences were D26, D54, and I143 in the AV40 strain and were G26, none, and T134 in the others. With respect to the SPV64 gene, the difference was L49 in AV40 and V49 in others (Figure 6).
Discussion
According to previous reports, outbreaks of sheep pox mainly occurred in Africa and middle Asia [4-7]. Capripox was widely epidemic in northwest, central, and southern China from 2000-2009 [20]. As a northern province in China, the information of outbreaks of sheep pox in the Shanxi area in recent years were incomplete. In the current study, a natural outbreak of the SPPV occurred in Shanxi province was reported. Diagnosis of the Capripoxvirus was initially based on clinical symptoms, followed by laboratory confirmation. In this research, the SPPV-Shanxi were identified by multiple methods. The CPE was inhibited by SPPV-specific antiserum. When cultured in LT cells, the viruses presented typical CPEs [21]. Electron microscopy showed the viruses were brickshaped, containing a central core which ruled out the involvement of Parapoxvirus infection because parapoxviruses are covered with a continuous filament [22,23]. The results of NT further confirmed that the disease outbreaks in Shanxi province were due to infection of the SPPV.
Based on alignment and phylogenetic analyses of the P32 gene, all strains were separated into three groups (Figure 3). In the first group, we found that the HM770955-Shanxi isolate (SPPV, China) and HQ607368-AV40 (SPPV, a Chinese SPPV vaccine strain) belonged to the same group. In addition, the SPPV and GTPV groups were all comprised of Chinese, and southern and central Asian strains. With respect to alignment of the amino acid residues, we found that the HM770955- Shanxi isolate, HQ607368-AV40, and FJ748487.1 (SPPV, India) have the same amino acid sequences, which revealed there might be a close relationship between the Shanxi isolate, Chinese SPPV vaccine AV40, and India isolate. In addition, we observed that all LSDVs (AF124516.1, AF32528.1, AF409137.1, and AF409138.1) represented a peculiar phenylalanine at position 49 instead of leucine, which exists in SPPVs and GTPVs. Moreover, we found that only FJ748488.1 (SPPV, India) had a glycine at the 26 position, which is the same as GTPVs. Whether or not glycine at position 49 is a signal of GTPV or it was derived from eastern or southern Asia might require more evidence.
Based on alignment and phylogenetic analyses of the SPV64 gene, all strains were divided into two groups (Figure 4). Although the Shanxi isolate HM802211 and AV40 strain HQ620556 were in separate groups, both had the same amino acid residues as SPPVs, GTPVs, and LSDVs. The remaining strain, AY077832 (SPPV, Turkey), had a different amino acid sequence and this finding might indicate that the SPPV-Shanxi isolate HM802211 was not derived from western Asia.
Hence, detection of the sequence of the P32 and SPV64 genes of the SPPV-Shanxi isolate revealed that the SPPV-Shanxi isolate is different from all previously published Capripoxvirus sequences. Our research may provide valuable epidemic evidence in Shanxi province and useful information on the genotype of SPPV-Shanxi. Furthermore, the efficiency of the PCR method in characterization of Capripoxvirus was confirmed. In addition, the outcomes might further assist us to explore the specificity of determinants of SPPV-Shanxi on the basis of molecular biology research.
Funding
This research was supported by the National Key Technologies R&D Program during the 11th Five-Year Plan period of China (2007BAD56B06).
Conflict of Interest
The authors declare that there is no conflict interest.
Authors’ Contributions
Shao-peng Gu conceived and coordinated the study, performed the histopathology, most of the gross pathology and drafted the preliminary manuscript; Xin-tao Shi, Ming-xue Zheng, Zongyong Shi, Jian-qin Yuan, Yun Li participated in the gross pathology and the presentation of data; Xin-tao Shi and Xingguo Liu participated in the literature research; Shao-peng Gu, Xin-tao Shi, Ming-xue Zheng, Zong-yong Shi and Jian-qin Yuan participated in the studies analysis; Shao-peng Gu, Xintao Shi, Nai-rui Huo and Rui Bai contributed to analysis and discussions all authors contributed to writing the draft manuscript and read and approved the final manuscript.
Acknowledgement
We thank Genetically Modified Testing Center, Ministry of Agriculture, China, for their help. We also thank the staff members of the Laboratory of Veterinary Pathology, Shanxi Agricultural University, China, for their help.
References
- Babiuk S, Bowden TR, Boyle DB, Wallace DB, Kitching RP. Capripoxviruses: an emerging worldwide threat to sheep goats and cattle. Transbound Emerg Dis 2008; 55: 263-272.
- Yeruham I, Yadin H, Van Ham M, Bumbarov V, Soham A, Perl S. Economic and epidemiological aspects of an outbreak of sheeppox in a dairy sheep flock. Vet Rec 2007; 160: 236-237.
- Bhanuprakash V, Indrani BK, Hosamani M Singh RK. The current status of sheep pox disease. Comp Immunol Microbiol Infect Dis 2006; 29: 27-60.
- Asagba MO, Nawathe DR. Evidence of sheep pox in Nigeria. Trop Anim Health Prod 1981; 13: 61.
- Achour HA, Bouguedour R. Epidemiology of sheep pox in Algeria. Rev Sci Tech 1999; 18: 606-617.
- Kitching RP, Bhat PP, Black DN. The characterization of African strains of capripoxvirus. Epidemiol Infect 1989; 102: 335-343.
- Mariner JC, House JA, Wilson TM, van den Ende M, Diallo I. Isolation of sheep pox virus from a lamb in Niger. Trop Anim Health Prod 1991; 23: 27-28.
- Kitching RP, McGrane JJ, Hammond JM, Miah AH, Mustafa AH. Capripox in Bangladesh. Trop Anim Health Prod 1987; 19: 203-208.
- Mondal B, Hosamani M, Dutta TK, Senthilkumar VS, Rathore R. An outbreak of sheep pox on a sheep breeding farm in Jammu, India. Rev Sci Tech 2004; 23: 943-949.
- Bhanuprakash V, Moorthy AR, Krishnappa G, Srinivasa Gowda RN, Indrani BK. An epidemiological study of sheep pox infection in Karnataka State, India. Rev Sci Tech 2005; 24: 909-920.
- Bowden TR, Coupar BE, Babiuk SL, White JR, Boyd V, Duch CJ, Shiell BJ, Ueda N, Parkyn GR, Copps JS, Boyle DB. Detection of antibodies specific for sheep pox and goat pox viruses using recombinant capripoxvirus antigens in an indirect enzyme-linked immunosorbent assay. J Virol Methods 2009; 161: 19-29.
- Beard PM, Sugar S, Bazarragchaa E, Gerelmaa U, Tserendorj SH, Tuppurainen E, Sodnomdarjaa R. A description of two outbreaks of capripoxvirus disease in Mongolia. Vet Microbiol 2010; 142: 427-431.
- Zeng XC, Chi XL, Li W, Hao WB, Li M, Huang XH, Huang YF, Rock DL, Luo SH, Wang SH. Complete genome sequence analysis of goatpox virus isolated from China shows high variation. Vet Microbiol 2014; 173: 38-49.
- Chand P. Molecular and immunological characterization of amajor envelope protein of capripoxvirus. Ph.D. Thesis University Surrey UK 1992; 227: 187-196.
- Townsley AC, Senkevich TG, Moss B. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J Virol 2005; 79: 10988-10998.
- Tulman ER, Afonso CL, Lu Z, Zsak L, Sur JH, Sandybaev NT, Kerembekova UZ, Zaitsev VL, Kutish GF, Rock DL. The genomes of sheeppox and goatpox viruses. J Virol 2002; 76: 6054-6061.
- Commission of Chinese Veterinary Pharmacopeia,Veterinary Pharmacopoeia of PRC Ministry of Chinese Agriculture press China 2010; 11: 3.
- Babiuk S, Parkyn G, Copps J, Larence JE, Sabara MI, Bowden TR, Boyle DB, Kitching RP. Evaluation of an ovine testis cell line (OA3.Ts) for propagation of capripoxvirus isolates and development of an immunostaining technique for viral plaque visualization. J Vet Diagn Invest 2007; 19: 486-491.
- Gallina L, Scagliarini L, McInnes CJ, Guercio A, Purpari G. Parapoxvirus in goats: experimental infection and genomic analysis. Vet Res Commun 2008; 32: 203-205.
- Zhao Z, Fan B, Wu G, Yan X, Li Y. Development of loop-mediated isothermal amplification assay for specific and rapid detection of differential goat pox virus and sheep pox virus. BMC Microbiol 2014; 14: 10.
- Situation of Capripox in China. Chinese Agr Sci Bull 2010; 26: 6-9.
- Rao SMV, Malik BS, Sharma SN. Antigenic relationship among sheep pox goat pox and contagious pustular dermatitis viruses. Acta Virol 1984; 28: 380-387.
- Kitching RP. Sheep pox and goat pox. Infectious Disease of Livestock Oxford University Press Southern Africa 2004; 2.