ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Rapid Communication - Biomedical Research (2019) Volume 30, Issue 1
Identification of breed-specific SNP panel in nine different cattle genomes.
Harshit Kumar1, Manjit Panigrahi1*, Supriya Chhotaray1, Dhan Pal1, V. Bhanuprakash1, K. A. Saravanan1, Rahul Shandilya1, Subhashree Parida2 and Bharat Bhushan1
1Division of Animal Genetics, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, UP, India
2Division of Pharmacology & Toxicology, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, UP, India
- *Corresponding Author:
- Manjit Panigrahi
Division of Animal Genetics
ICAR-Indian Veterinary Research Institute, India
Accepted date: December 28, 2018
DOI: 10.35841/biomedicalresearch.30-18-1195
Visit for more related articles at Biomedical ResearchSingle nucleotide polymorphism (SNP) high-density chips are now serving as important bioinformatics tools for improvement and development of various livestock species. Major constraint being the high cost of protocol which is not feasible at the population level. Hence, in the present study, we have tried to reduce the SNP panel to a fewer number of informative markers which will be very much cost-effective. The 50K Illumina BeadChip genotypic data obtained from online Dryad repository for six indigenous cattle breeds, namely Tharparkar, Hariana, Red Sindhi, Sahiwal, Gir and Kankrej were merged with three exotic breeds mainly used in Indian condition, i.e., Holstein-Friesian, Jersey and Brown Swiss. Various quality parameters (MAF-0.36, hwe-0.001, geno-0.95) and statistical operations (FST, LD values) were applied by different bioinformatics tools. Later, best possible SNPs with an average FST value of >0.8 were analysed using STRUCTURE 2.3.4 software and we have found perfect clustering among the nine breeds comprising a total of 536 SNPs referring to 158 individuals from nine breeds. Later, breed-specific SNPs were filtered from the set of 536 SNPs using Venny 2.1.0 software.
Keywords
Informative markers, Indigenous cattle, Linkage disequilibrium, Minor allele frequency, SNPs
Introduction
Indigenous cattle breeds are well adapted to our agro-climatic conditions and are resistant to many tropical diseases. It can survive and produce milk on poor feed and fodder resources. Some of these breeds are well established for their high milk and fat production. However, the production potential of these animals has deteriorated over a period due to lack of selection [1]. The high producing exotic breeds do not have the above characteristics and are very difficult to manage in the tropical Indian scenario. Hence, indigenous cattle breeds should be improved and conserved at their breeding tracts.
One approach is to identify the purebred animals, with the advent of high-density genotyping of blood samples and rapid availability of Bovine50K and HD SNP data. Bovine SNP high-density chips are useful but the cost of operations would be much higher. Hence, there is a need for a cost-effective protocol which is possible by identifying the small number of highly informative SNPs [2]. Several studies have shown the implications of SNPs in differentiating breeds of individuals in the population and also assigning an individual to its population of origin [3-5]. Further, the protocols to filter and select highly informative markers which makes differentiation at the breed level and assigning of individuals to its specific breeds have been described in several reports [3,6-8].
Breed-specific SNPs were identified using Reynolds FST and extended Lewontin and Krakauer's (FLK) statistics by Zwane and his co-workers among three South African indigenous breeds after filtering them at 0.05 MAF [9]. Recently, the genetic diversity among three indigenous dairy cattle breeds of India, viz., Sahiwal, Tharparkar, and Gir were analysed based on BovineHD SNP data. Fifty percent of the SNPs of this assortment were found to be informative for genetic analysis of these cattle. The common SNPs with MAF ranging from 0.1 to 0.5 were approximately 50% and 34% for BovineHD and 54K Chips, respectively [10]. In another report, only SNPs in Hardy-Weinberg equilibrium, displaying the highest Minor Allele Frequency across all the thirty populations of French sheep breed (not associated with Mendelian errors in verified family trios) were selected. A panel of 249 SNPs was successfully used in an on-farm test in the BMC breed (Blanche du Massif Central) sheep and resulted in more than 95% of lambs being assigned to a unique sire [11]. Therefore, multiple level filtrations of SNPs has been attempted to cut the number of SNPs at various levels. Yousefi et al. [12] obtained various subsets via routine filtering of markers by taking into consideration the minor allele frequency, genotype call rate, missing rate of individuals to produce high-quality subsets from crude SNPs. The data were further exposed to restrictive filtering with significant levels of Hardy-Weinberg equilibrium and linkage disequilibrium (LD) into consideration to obtain SNP panel of 50 markers for individual assignment.
Hence, in the current study, we attempted a different approach to reduce the number of SNPs from Bovine50K chip data of nine breeds of cattle, i.e., six indigenous cattle along with three exotic breeds (commonly used in India), available online at Dryad repository. The reduced SNP panel will be helpful to identify individuals of a particular breed in a cost-effective manner and further to augment various breeding strategies for improvement of indigenous cattle breeds in India.
Material and Methods
Preparation of preliminary dataset
To prepare the dataset we obtained the genotypic data from four high yielding indigenous milch cattle breeds, i.e., Tharparkar (12), Red Sindhi (10), Sahiwal (17) and Gir (24) with two dual-purpose breeds Hariana (10) and Kankrej (10) from Dryad repository (13) data for. ped/.map files accessed via WIDDE (Web-Interfaced next generation database for genetic diversity exploration). Three exotic cattle breeds had been extensively used in India in the past six decades for cross-breeding programmes. Hence, in the present study we have also taken three exotic breed’s genotypic data, i.e., Holstein Friesian (30), Jersey (21) and Brown Swiss (24) along with the above-said files. Finally, a nine datasets comprising a total of 158 individuals were obtained and subjected to further quality control parameters. All the animals obtained from online repository were genotyped using Illumina BovineSNP50v2 BeadChip [13].
Quality control, filtering and selection of SNPs
Genotype and major/minor allele frequencies were then calculated using PLINK [14]. Minor allele frequency was calculated based on the frequency of the least common allele for every SNP in the given population [12]. We carried out filtering of SNPs within individual breed files as per following criteria, i.e., (a) SNPs with genotype calling rate less than 95%, (b) SNPs with more than three genotype and more than two alleles, (c) SNPs with minor allele frequency less than 0.36. Afterwards, each dataset was subjected to Hardy-Weinberg equilibrium filter at 0.001 statistic followed by pruning SNPs with pairwise LD. The LD has been defined as the non-random relationship between alleles at diverse loci within a population. It was performed by taking a window of 50 SNPs and removing a pair which have calculated value of LD greater than 0.01 (r2). Later, the window was shifted by five SNPs forward and repeating the procedure for all the nine datasets, which generated pruned subset of SNPs which were in approximate linkage equilibrium with each other based on pairwise genotypic correlation.
LD based reduction of SNPs
All the LD pruned nine datasets were further merged using PLINK software. To obtain the final panel, SNPs were subjected to pruning again based on pairwise genotypic correlation method taking similar parameters as discussed above. The final dataset was subjected to genetic analysis using STRUCTURE software. In STRUCTURE, data were subjected to 20,000 burn-in and 30,000 MCMC runs for all the 10 iterations. Further, FST values were inferenced from STRUCTURE analysis to designate the SNPs as informative [9]. A higher FST value for any SNP suggest that a high level of variation for that SNP has occurred in members of the subpopulation equated to the total population, and thus members of the subpopulation incline to carry distinctive informative alleles compared to the total population [15].
Result and Discussion
An SNP was declared to be breed-specific when it possessed an allele that was present in only one breed [7]. Numerous studies have proved the usefulness of SNP data for identifying breed informative SNPs for genetic discrimination of breeds [3,6]. After applying the above mentioned quality parameters, i.e., MAF, Hardy-Weinberg equilibrium, genotype call rate and LD pairwise pruning we obtained nine sets comprising of a total of 1324 informative SNPs (Table 1). After merging the nine dataset, the final dataset was further pruned again via pairwise LD (r2=0.01), to obtain a set of 536 SNPs. Hence, to prune breed-specific SNPs in our effort, 1324 markers (nine SNP lists) were taken and compared with the final list of 536 SNPs using VENNY 2.1.0. The SNPs for specific breed were obtained which were not present in any other breeds. Such list of breed-specific SNPs was extracted one by one for all the breeds. We obtained a total of 470 breed-specific SNPs excluding 66 SNPs which were in common in one or the other breed (Table 2). As shown in Tables 1 and 2 we were able to reduce SNP marker set of 53,074 from Bovine50 BeadChip to 470 SNPs in our trial. Yousefi et al. could also reduce panel to 50 SNPs using similar quality parameters for human DNA/RNA identification [12].
| Cattle Breed | No of SNPs |
|---|---|
| Tharparkar | 100 |
| Sahiwal | 88 |
| Red Sindhi | 120 |
| Gir | 128 |
| Hariana | 92 |
| Kankrej | 132 |
| Holstein Friesian | 254 |
| Jersey | 227 |
| Brown Swiss | 183 |
Table 1: Details number of SNPs obtained from individual breeds after applying quality parameters.
| Cattle Breed | No of SNPs |
|---|---|
| Tharparkar | 43 |
| Sahiwal | 39 |
| Red Sindhi | 47 |
| Gir | 32 |
| Hariana | 36 |
| Kankrej | 40 |
| Holstein Friesian | 77 |
| Jersey | 87 |
| Brown Swiss | 69 |
Table 2: Breed specific SNPs obtained using Venny 2.1.0.
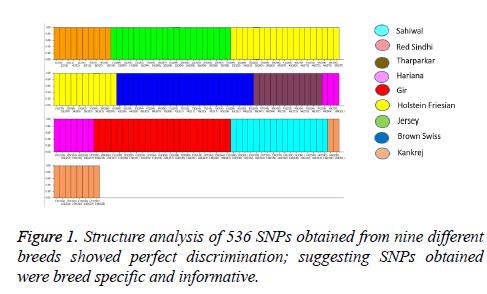
Structure analysis (K=9) was performed to evaluate the genetic structure and affinities among the nine populations included in our study. Figure 1 illustrates the clustering of the different breeds, showing the perfect discrimination of six indigenous cattle breeds (Tharparkar, Hariana, Red Sindhi, Sahiwal, Gir and Kankrej) and three exotic breeds (Holstein Friesian, Jersey and Brown Swiss). The cluster of the nine breeds showed that the observed pattern of clustering separated these populations based on their genotyping platforms, i.e., Bovine SNP50. These structure results, as expected, show that there is perfect discrimination of nine breeds based on our reduced SNP panel of 536 SNPs. Makina et al. performed similar genetic differentiation among six South African cattle breeds using structure analysis [16].
The informative SNPs having high wrights FST values may further be identified on different chromosomes. In the present study we obtained an average of FST values above 0.8. In addition, we would also like to mention that all previous studies and data analysed for finding informative SNPs widely used bi-allelic data, but still many reports are available for triallelic markers being highly informative. Many human identification panels have been developed based on tri-allelic SNPs [17] as tri-allelic SNPs have more discriminatory power [18]. Recently, 8 tri-allelic SNPs were introduced in panel for biogeographical ancestry identification among Chinese Han population [19]. Hence, we further suggest in future introducing informative tri-allelic SNPs studies may bring even better and precise determination of breed purity.
The analyses performed in our study were conducted to identify breed informative markers panel for use in discriminating indigenous cattle breeds by using the BovineSNP50 data [13]. Although the Bovine SNP50 assays were designed to contain variants that were common to taurine breeds, the authors concluded the usefulness of established methodology in identifying informative SNPs to discriminate different indigenous cattle breeds.
Acknowledgement
The authors would wish to acknowledge Director, IVRIBareilly, for their support and providing infrastructural facility to conduct this study.
Data Availability Statement
Genotypic data used for our analysis can be found at WIDDE database or at Dryad repository. Links are provided herein; http://widde.toulouse.inra.fr/widde/widde/main.do? module=cattle and https://doi.org/10.5061/dryad.th092 , respectively.
Disclosure Statement
The authors declare no conflict of interest.
References
- Lewis J, Abas Z, Dadousis C, Lykidis D. Tracing cattle breeds with principal components analysis ancestry informative SNPs. PloS One 2011; 6: 18007.
- Matukumalli LK, Lawley CT, Schnabel RD. Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 2009; 4: 5350.
- Wilkinson S, Wiener P, Archibald AL. Evaluation of approaches for identifying population informative markers from high density SNP chip. BMC Genetics 2009; 12, 1-14.
- Dimauro C, Cellesi M, Steri R. Use of the canonical discriminant analysis to select SNP markers for bovine breed assignment and traceability purposes. Animal Genetics 2013; 44: 377-382
- Hulsegge B, Calus MPL, Windig JJ. Selection of SNP from 50K and 777K arrays to predict breed of origin in cattle. J Animal Sci 2013; 91: 5128-5134.
- Negrini R, Nicoloso L, Crepaldi P. Assessing SNP markers for assigning individuals to cattle populations. Animal Genetics 2009; 40: 18-26.
- Ramos AM, Megens HJ, Crooijmans RPMA. Identification of high utility SNPs for population assignment and traceability purposes in the pig using high‐throughput sequencing. Animal Genetics 2011; 42: 613-620.
- Opara A, Razpet A, Logar B. Breed assignment test of Slovenian cattle breeds using microsatellites. Acta Agric Slov 2012; 3: 167-170.
- Zwane AA, Maiwashe A, Makgahlela ML. Genome-wide identification of breed-informative single-nucleotide polymorphisms in three South African indigenous cattle breeds. South African J Animal Sci 2016; 46: 56.
- Dash S, Singh A, Bhatia AK. Evaluation of bovine high-density SNP genotyping array in indigenous dairy cattle breeds. Animal Biotechnol 2017; 20: 1-7.
- Tortereau F, Moreno CR, Tosser-Klopp G. Development of a SNP panel dedicated to parentage assignment in French sheep populations. BMC Genetics 2017; 18: 50.
- Yousefi S, Abbassi-Daloii T, Kraaijenbrink T. A SNP panel for identification of DNA and RNA specimens. BMC Genomics 2018; 19: 90.
- Decker JE, McKay SD. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genetics 2014; 10: 1004254.
- Purcell S, Neale B, Todd-Brown K. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Human Gene 2007; 81.
- Norrgard K, Schultz J. Using SNP data to examine human phenotypic differences. Nat Educ 2008; 1: 85.
- Makina SO, Muchadeyi FC, Van Marle-Koster E. Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel. Front Gene 2014; 5: 333.
- Zha L, Yun L, Chen P. Exploring of tri-allelic SNPs using pyrosequencing and the SNaPshot methods for forensic application. Electrophoresis 2012; 33: 841-848.
- Western D, Russell S, Cuthill I. The status of wildlife in protected areas compared to non-protected areas of Kenya. PLoS One 2009; 4: 6140.
- Gao Z, Chen X, Zhao Y. Forensic genetic informativeness of an SNP panel consisting of 19 multi-allelic SNPs. Forensic Sci Int Genet 2018; 34: 49-56.