ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Review Article - Biomedical Research (2015) Applications of Rapid Prototyping Techniques in Bio-Materials ARTBM2015
In silico approach of antibacterial compounds from mangrove - Avicennia marina through docking analysis.
aAssistant Professor, Department of Biotechnology, Karpaga Vinayaga College of Engineering and Technology, Chinna Kolambakkam, Tamilnadu, India
bProfessor, Department of Biotechnology, Rajalakshmi Engineering College, Tamilnadu, India
- *CorresponSding Author:
- A. Sheela Devi
Department of Biotechnology
Karpaga vinayaga college of engineering and technology,
Chinna Kolambakkam, Tamilnadu India
Accepted July 07 2015
The term “In - silico” mean “any biological experiment on or in computer”. This method is easy, relatively fast and used to predict pharmacology and/or toxicology hypothesis. In - silico software have been employed in the discovery and optimization of novel bioactive compounds with affinity to a particular target, the clarification of absorption, distribution, metabolism, excretion and toxicity properties (ADME/Tox) as well as physicochemical characterization. Urinary tract infection (UTI) is a condition in which the urinary tract is infected with a patho-gen causing inflammation which is a widespread, difficult and rarely life threatening condition. Avicennia marina contains biologically active antibacterial, antiviral and antifungal compounds. Hence the antibacterial com-pounds of A. marina such as beta humulene, taraxasterol, alpha amyrin, venlafaxine, 3-(3-methoxy phenyl) pro-panoic acid and methyl 2-methylinden-3-on carboxylate were selected for this study. The docking was performed between ligands and virulent proteins of UTI pathogens, thus LIBDOCK score were analyzed, followed by Lipinski’s rule of 5 were calculated. Drug like properties of these ligands were calculated by ADME calculations. Based on these results, out of 6 compounds 2 were confirmed as promising compounds against the particular pathogens. Therefore the present study play a guiding role to develop new inhibitors with better binding affinities towards the specific virulent protein of pathogens, followed by invention of novel drug to treat UTI.
Keywords
Avicennia marina, drug discovery, mangrove plant, molecular docking, urinary tract infection
Introduction
Antimicrobial properties of medicinal plants are being increasingly reported from different parts of the world [1]. Plants are very significant to human, because they have several dynamic constituents which are precursor to syn-thesize many drug [2]. India has a rich flora that is widely distributed throughout the country. Herbal medicines are basis of treatment and cure variety of diseases. Plant based drugs were commonly used in India and China [3]. Mangrove plants and their products have been widely used in traditional medicine. These plants have various natural products with enormous pharmaceutical signifi-cance and also exhibiting antimicrobial, anti viral, anti larval and anti insecticidal activity [4]. Urinary tract is among the most common sites of bacterial infection in humans [5]. Urinary tract infections (UTIs) are among the most prevalent infectious diseases, with a extensive eco-nomic burden on society [6]. Escherichia coli is the commonest cause of all type of UTIs [7]. UTI was more widespread among females compared to males. E. coli was the most prevalent isolates (43.9%), followed by Staphylococcus aureus (21.6%), Candida albicans (16.4%), Klebsiella pneumonia (14.7%) and Pseudomo-nase aeruginosa (3.4%) [8]. Cytotoxic necrotizing factor 1 (CNF1) are toxins produced by uropathogenic E.coli (UPEC). CNF1 exerted a strong positive influence on expression of genes involved in innate immunity and sig-nal transduction and a negative impact on metabolism and transport associated genes [9]. It is 115-kDa and ex-pressed by 40% of UPEC isolates and up to 30% of diar-rheal E. coli isolates [10]. Lipopolysaccharides are neces-sary for the activity of the protease (OmpT) [11]. OmpT is one of the virulent genes thought to be important in E. coli mediated UTI [12]. Alpha-toxin known as alpha-hemolysin (Hla), is the major cytotoxic agent released by S. aureus [13]. This toxin consists chiefly of beta-sheets (68%) with only about 10% alpha-helices, and it perform major function in host cell, that is development of pores in the cellular membrane, eventually causing cell death [14]. Staphylococcal protein A (SPA) is an important virulence factor which enables S. aureus to evade host immune responses [15].
Methodology
Selection of protein and preparation of its structure
Rho activating domain of E. coli CNF1 (PDB ID: 1HQ0), crystal structure of ompT from E. coli (PDB ID: 1I78), SPA (PDB ID: 2JWD) and alpha-hemolysin from S. au-reus (PDB ID: 7AHL) were selected in our study. The protein structures were downloaded from protein data bank (http://www.rcsb.org/pdb/) establish ed by Brookhaven National Laboratories (BNL) in 1971. The proteins were prepared for docking by the removal of wa-ter molecules and heteroatom from the downloaded pro-tein structures. Crystallographic disorders and unfilled valence atoms were corrected using alternate confor-mations and valence monitor options and were subjected to energy minimization by applying CHARMM (Chemis-try at HARvard Macromolecular Mechanics) force fields. CHARMM is program for macromolecular dynamics; it can be used for energy minimization, normal modes and crystal optimizations and also X-Ray incorporates free energy methods for chemical and conformational free calculations.
Selection of ligand and preparation of its structure
Beta humulene (L1), taraxasterol (L2), alpha amyrin (L3), venlafaxine (L4), 3-(3-methoxy phenyl) propanoic acid (L5) and methyl 2-methylinden-3-on carboxylate (L6) present in A.marina were selected in our study. The struc-tures of the ligands were downloaded from PubChem da-tabase. Hydrogen bonds were added and the energy was minimized using CHARMm force field.
Interaction studies of binding and calculation of ADME / TOX
The exact fit of the ligand to a receptor was studied using LibDock module in the Discovery Stu-dio Accelrys® software corporation, San Diego, USA. The interactions of the drugs with the target were ana-lyzed using the receptor-ligand interaction protocol of the software. The receptor cavities were explored and the active site residues selected were used for the interaction with the drugs. With the intention of find out the drug like properties for the ligands ADME calculation was per-formed by using Accord for Excel 6.1, an accelry’s prod-uct.
Results and Discussion
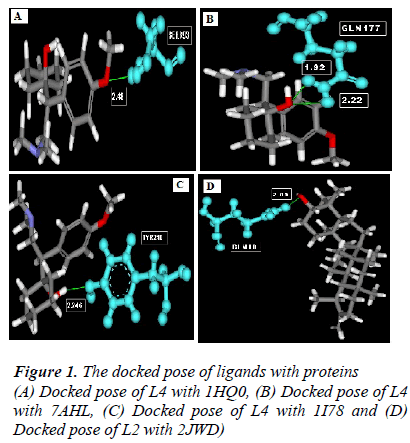
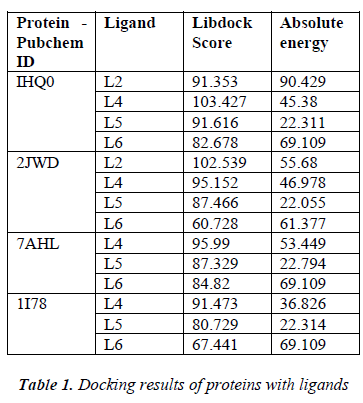
According to the result of docking, out of 6 compounds, only 4 had docking with Rho activating domain of E. coli CNF1 and SPA; and 3 had docking with alpha-hemolysin and ompT. It was given in table 1. The compound ven lafaxine had high Libdock score with Rho activating do-main of E. coli CNF1, alpha hemolysin and ompT. Com-pound taraxasterol had high score with SPA. The docked pose of these 4 were shown in Fig 1 (A, B, C and D re-spectively). In Fig 1, the green lines denoted the hydrogen bonds. All the amino acid residues which involved in mo-lecular interactions were shown in Blue color and the Ligands were shown in grey color. ADME prediction is the most crucial step in drug discovery. Drugs which sat-isfy these properties only will survive in the Phase 1 clin-ical trial [16]. Out of 6, most of the compounds satisfy ADME properties and Lipinski’s rule of 5.
Based on the in silico drug designing analysis through molecular docking results and ADME values, the two active principles such as venlafaxine and taraxasterol from A.marina were confirmed as a promising antibacte-rial compound against E. coli and S. aureus. Hence fur-ther analysis through invitro and invivo studies will prove the action of these compounds.
Conclusion
Thus we concluded that the compounds render antimicro-bial action against UTI pathogens, hence it may be used as drug to treat UTI. The results of our study not only give a base for further research but also useful for drug development. Hence our study should therefore play a guiding role in the experimental design and development of antibacterial drug in the present and future to treat UTI and many ailments. Also our study provides us hope to overcome failures of drug resistance profiles. This study also means a natural alternative to antibiotics, which is an exhilarating and potentially extreme area of research.
Acknowledgment
We would like to thank Prof. Dr. Niranjali Devaraj, De-partment of Biochemistry, Chennai University and Prof. Dr. Ramalingam, Department of Biotechnology, Anna University for their valuable suggestions.
References
- Rai PK, Mehta S, Gupta RK, Watal G. “A Nov- el Antimicrobial Agents Trichosanthes Diocia” Inter- national Journal of Pharma and Bioscience 2010; 1(3): 1-9.
- Dhankhar Sandeep, Ruhil S. “Aegle marmelos Correa: A Potential Source of Phytomedicine” Jour- nal of Medicinal plant Research 2011; 5(9): 1497–1507.
- Duraipandiyan V, Ignacimuthu S. “Antibacteri- al and antifungal activity of Cassia fistula L. : an eth- no medicinal plant” J. Ethnopharmacol 2007; 112(3): 590-594.
- Premnathan M, Chandra K, Bajpai S.K, Kathiresan K. “A survey of some Indian marine plants for antiviral activity” Botanica marina 1992; 35(4): 321-324.
- Kunin CM. “Urinary tract infections in females”Clin Infect Dis1994; 18(1):1-12.
- Grabe M et al “Guidelines on Urological Infec- tions.” European Association of Urology, 2011.
- Leslie C, Albert B, Sussman M. “Urinary Tract Infections” Microbiology and microbial infections 1998; 9(3): 1005-1009.
- Al-Jiffri O et al “Urinary Tract Infection with Esherichia coli and Antibacterial activity of some plants extracts” International Journal of Microbiologi- cal Research 2011; 2(1): 01-07.
- Tamako, Garcia and Christy A, “Ventura L.Cytotoxic Necrotizing Factor 1 and Hemolysin from Uropathogenic Escherichia coli Elicit Different Host Responses in the Murine Bladder” Infection and Im- munity 2013; 81 (1): 99–109.
- Hofman P et al “Rossi B. Escherichia coli cyto- toxic necrotizing factor-1 (CNF-1) increases the ad- herence to epithelia and the oxidative burst of hu- man polymorphonuclear leukocytes but decreases bacteria phagocytosis” J. Leukoc. Biol 2000; 68(4): 522–528.
- Kramer RA et al “Lipopolysaccharide regions involved in the activation of Escherichia coli outer membrane protease OmpT” Eur. J. Biochem 2002; 269(6): 1746– 1752.
- Carl F. Marrs et al “Variations in 10 putative uropathogen virulence genes among urinary, faecal and per-urethral E. coli ” J. Med. Microbiology 2002; 51(2):138-142.
- Bhakdi S, Tranum-Jensen. “α-toxin of Staphylococcus aureus” Microbiol Rev 1991; 55(4): 733–51.
- Bubeck Wardenburg J, Schneewind O. “Vac- cine protection against Staphylococcus aureus pneumonia” J. Exp Med 2008; 205(2): 287–94.
- Antonina A et al, “Prevalence of Staphylo-coccus aureus protein A (spa) mutants in the com- munity and hospitals in Oxfordshire” BMC Micro- biology 2014; 14(63): 1-11.
- Lipinski CA, Lombardo F, Dominy Bw, Feeney PJ. “Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings” Adv Drug Deliv Rev2001; 46: 3-25.