ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2016) Volume 27, Issue 4
Investigating multidrug efflux pumps in relation to the antibiotic resistance pattern in Escherichia coli strains from patients in Iran
Farshid Kafilzadeh1* and Farnoosh Farsimadan2
1Department of Biology, Jahrom Branch, Islamic Azad University, Jahrom, Iran
2Department of Biology, Kazeroon Branch, Islamic Azad University, Kazeroon, Iran
- *Corresponding Author:
- Farshid Kafilzadeh
Department of Biology Jahrom Branch
Islamic Azad University Jahrom
Iran
Accepted date: March 31, 2016
Objective: To investigate antibiotic resistance pattern of Escherichia coli strains isolated from patients admitted to the Shahid Mostafa Khomeini hospital in Behbahan city and survey its relationship with the presence of efflux pumps.
Methods: In this study, 180 isolates of E. coli were collected from Shahid Mostafa Khomeini hospital. After confirmation of strains by standard culturing methods and biochemical tests, their antibiotic resistance was evaluated with agar diffusion test and CLSI standards guidelines. The genes encoding the AcrA-AcrB-TolC efflux pump were identified with PCR.
Results: The percentages of resistance of the E. coli isolates to some antibiotics were as follow: novobiocin and rifampin, 68.9%; erythromycin, 52.8%; tetracycline, 52.2% and nalidixic acid, 51.7%. Isolates were sensitive to amikacin, 89.4%; tobramycin, 85.6%; meropenem, 81.7%; chloramphenicol, 77.8% and piperacillin, 65.6%, respectively. 110 (61.1%) strains from 180 taken isolates were found with multidrug- resistance (MDR) but no pandrug-resistance (PDR) was observed. 51.1%, 75.0% and 69.4% of isolates had genes acrA, acrB and tolC, respectively. Comparison of the results of antibiogram obtained from PCR with the two-way correlation test showed that there is a significant positive correlation between the presence of efflux pumps and resistance to antibiotics (excluding carbenicillin, meropenem, chloramphenicol, cefotaxime, rifampin and novobiocin).
Conclusion: Due to differences in antibiotic resistance in different strains is required using antibiotic resistance pattern for experimental and specific treatment of patients.
Keywords
Multi-drug resistance, E. coli, Antibiotic resistance, Efflux pumps, Behbahan.
Introduction
Significant increases in prevalence of resistance to antibiotics in different populations of humans and animals pathogens are among of the most serious global public health threats. Overuse of antibiotics is the most frequent cause of antibiotic resistance in pathogenic bacteria. The emergence of resistant organisms to antibiotic treatment is a global problem in societies and hospitals. Nowadays trend of response to the standard therapy of hospital infections has changed and the prevalence of antibiotic resistance in many hospitals reached dangerous levels. According to studies, about 50 to 60 percent of hospital infections are caused by antimicrobial-resistant strains [1,2]. Antibiotics are used in medicine to prevent, treat or reducing the incidence of infectious diseases in farm animals and human populations. Escherichia coli strains are the main cause of several common bacterial infections. This bacterium is rod-shaped, Gram-negative and facultative anaerobe that frequently in large numbers are the normal floras of humans gut. E. coli known as an opportunistic pathogen in patients with immune system disorder, which can cause serious infections in these patients. This bacterium is agent of urinary tract infection (UTI), respiratory tract infections meningitis, septicemia and gastroenteritis. Most E. coli infections except neonatal meningitis and gastroenteritis are endogenous [3,4]. Today, drug resistance especially among hospital patients has become the major clinical significance of E.coli. In addition E. coli is intrinsically resistant to particular antibiotics; this bacterium easily becomes drug resistant during treatment and this arise difficulties in the treatment of patients [5]. Nowadays the active efflux pumps have been considered as one of the most important intrinsic and acquired antibiotics resistance mechanisms in bacteria. Efflux pump not only increases the Minimum Inhibitory Concentration (MIC) against antibiotics but also creates mutant strains of antibiotics resistant bacteria with decrease in drug concentration inside the microbial cells. E. coli has been found to possess a variety of inducible tripartite drug efflux complexes such as AcrAB-TolC [6,7]. Nikaido and Zgurskaya demonstrated that the AcrB system of E. coli is a RND family multidrug efflux system that pumps out many amphiphilic and lipophilic inhibitors through the TolC outer membrane channel. AcrA, an elongated protein, brings both the inner and the outer membrane closer together. This protein acts as a trimmer and is in contrast to AcrB [8]. Antimicrobial resistance in E. coli have been reported around the world [9-11] and the increasing rate of resistance in this bacteria has established a lot of concern in developed and developing countries and therefore in order to avoid treatment failure and infection control antibiogram analysis should be performed before prescription of antibiotics to prevent the indiscriminate use. Given the lack of knowledge about the infection prevalence and antibiotic susceptibility patterns of E. coli in the Behbahan city, this study was carried out to determine the prevalence of infections and antibiotic susceptibility patterns of E. coli in Shahid Mostafa Khomeini Hospital and designate the relationship between presence of efflux pumps in E. coli and resistance to antibiotics and it is hoped that the results can be used in proper choice of effective and appropriate antibiotics for typical patients with infections.
Materials and Methods
Samples collection
In this study, a large number of different samples (wound, urine, blood) that was probably contaminated with E. coli were collected from admitted patients to Shahid Mostafa Khomeini Hospital, in Behbahan city within four months from February untill May, 2014. The written consent and authorization of the research ethics committee (2014-1-65-10433) and written informed consent of the patients were obtained in a questionnaire and then taking the samples was performed. Obtained samples were transported to the laboratory in the shortest possible time and bacterial culture was done. Finally, 180 samples were found contaminated with E.coli.
Identification of isolates
The isolates were identified by Gram staining and biochemical tests including oxidase, catalase, Simmons citrate agar, triple sugar iron (TSI) agar, Phenylalanine Deaminase (PD), blood agar, Eosin Methylene Blue (EMB) agar, MacConkey agar, Oxidation Fermentation (OF), Sulfide Indole Motility (SIM) medium and Methyl Red-Voges-Proskauer (MR-VP) broth (all chemicals were purchased from Merck, Germany).
Antibiogram tests
To determine the sensitivity of the isolated E. coli bacteria against antibiotics, antibiotic resistance patterns of clinical samples of patients with hospital acquired infection were determined by disc diffusion method, according to CLSI standards. For this purpose, single colonies of microbial isolates from a 24 h culture were transferred to physiological serum with sterile inoculating loop. After vortex mixing, turbidity of the solutions was compared visually with 0.5 McFarland turbidity. With reaching this turbidity level, samples were spread on the surface of Mueller-Hinton agar with spread plate method by sterile swabs. After a few minutes, disks were placed at an appropriate distance and finally the plates were incubated at 37°C. The results were obtained by measuring the diameter of the zones of inhibition and comparing with the last available tables after 18 hours. Used antibiotics in this study, were include: cefuroxime (30 μg), amikacin (30 μg), tobramycin (10 μg), rifampin (5 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), carbenicillin (100 μg), ceftazidime (30 μg), ticarcillin (75 μg), cefotaxime (30 μg), piperacillin (100 μg), cefepime (30 μg), erythromycin (15 μg), tetracycline (30 μg), chloramphenicol (30 μg), cloxacillin (1 μg), norfloxacin (10 μg) and meropenem (10 μg). All antibiotics were prepared from Patan-Teb Company, Iran. In this study, the reference strain E. coli (ATCC 25922) was used as a positive control to confirm the results of antibiotic susceptibility testing.
PCR test
The presence of efflux pump genes in isolated samples was investigated using Polymerase Chain Reaction (PCR) assay. In the first step, boiling methods was used to extract genomic DNA. Proliferation of acrAB and tolC genes in isolated E. coli was performed with three separate primer pairs (Table 1).
| acrA | F | 5’- CTCTCAGGCAGCTTAGCCCTAA |
| R | 5’-TGCAGAGGTTCAGTTTTGACTGTT | |
| acrB | F | 5’- GGTCGATTCCGTTCTCCGTTA |
| R | 5’- CTACCTGGAAGTAAACGTCATTGGT | |
| tolC | F | 5’ - AAGCCGAAAAACGCAACCT |
| R | 5’- CAGAGTCGGTAAGTGACCATC |
Table 1. The primers used for PCR.
Extracted DNA containing acrAB genes from bacterium (ATCC 25922) were used as a positive control in PCR assay. PCR conditions were as follows: 1 μl (10 mM) dNTPs mix, 1.5 μl (50 mm) MgCl2 solution, 5 μl DNA and 2.5 units of Taq DNA Polymerase (materials were purchased from CinnaGen Company, Iran), 20 pmol of each forward and reverse primers in a final volume of 50 μl. Proliferation of target gene was conducted with hot-start at 94°C for 5 minutes, then 35 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 1 min and 72°C for 5 min. PCR products were finally analyzed by electrophoresis on 1% agarose gel [12,13]. In the end, the relationships between variables were analyzed with chi-square test at significance level of 5%. All analyzes were done with SPSS version 18 (IBM SPSS Statistics).
Results
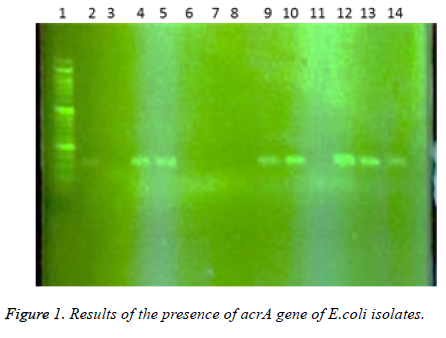
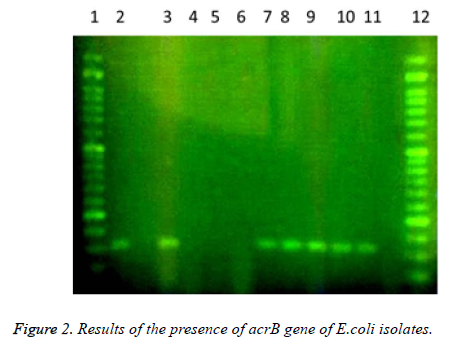
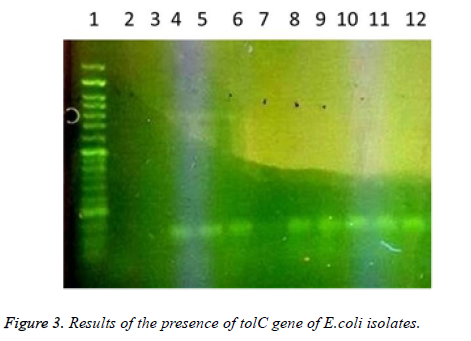
In this study a total of 180 samples of urine, blood and wounds contaminated with the E.coli were collected from patients admitted to the Shahid Mostafa Khomeini Hospital in Behbahan city. Isolated E.coli were verified with pink colonies on MacConkey agar and dark blue-black colonies with metallic green sheen on EMB agar and biochemical verification tests. In this study the presence of efflux pump genes were investigated using three separate primer pairs. Results are shown in Table 2 and Figures 1-3.
| Gene presence | ||||||
|---|---|---|---|---|---|---|
| Gene type | Positive | Negative | Total | |||
| absolute | Relative percentage | absolute | Relative percentage | absolute | Relative percentage | |
| acrA | 92 | 51.1 | 88 | 49.9 | 180 | 100 |
| acrB | 135 | 75 | 45 | 25 | 180 | 100 |
| tolC | 125 | 69.4 | 55 | 30.6 | 180 | 100 |
| Total | - | 100 | - | 100 | - | - |
Table 2. Results of presence of acrA, acrB and tolC genes in E. coli isolates.
1- 50 bp DNA ladder. 2- Samples with No. 47 and 148 with primers of acrA gene (positive). 3- Sample 32 with primers of acrA gene (negative and sensitive to all used antibiotics). 4,5- Standard reference strain of E. coli with primers of acrA gene. 6,7 and 8- Samples with No. 71, 63 and 54 with primers of acrA gene (negative). 9, 10- Samples with No. 80 and 112 with primers of acrA gene. 11- Sample 98 with primers of acrA gene (negative). 12, 13 and 14- Samples with No. 112, 49 and 1- 50 bp DNA ladder. 2,3- Samples with No. 9 and 148 with primers of tolC gene (negative). 4-Standard reference strain of E. coli with primers of tolC gene. 5, 6- Samples with No. 134 and 177 with primers of tolC gene (positive). 7- Sample 32 with primers of tolC gene (negative and sensitive to all used antibiotics). 8, 9, 10, 11 and 12- Samples with No. 31, 4, 121, 101 and 100 with primers of tolC gene (positive). In the following, the results of antibiotic susceptibility test showed that the highest sensitivity against amikacin (89.4%), tobramycin (85.6%), meropenem (81.7%), chloramphenicol (77.8%), piperacillin (65.6%), cefepime (66.1%), norfloxacin (63.3%), ciprofloxacin (59.4%) ticarcillin (57.8%), carbenicillin (52.8%), ceftazidime (54.4%), cefotaxime (52.2%), ticarcillin and nalidixic acid (46.7%), cefuroxime (36.1%), erythromycin (35.6%), novobiocin and rifampin (31.1%). The highest resistance were observed against novobiocin and rifampin (68.9 %), erythromycin (52.8%), tetracycline (52.2%), nalidixic acid (51.7%), cefuroxime (47.8%), carbenicillin (42.8%), cefotaxime (42.2%), ticarcillin (40.6%), ceftazidime (38.9%), norfloxacin (32.2%), ciprofloxacin (31.1%), cefepime (30.6%), piperacillin (17.2%), chloramphenicol (14.4%), meropenem (12.2%), tobramycin (10.6%) and amikacin (8.7%), respectively (Table 3).
| Antibiotic | Resistant (R) | Sensitive (S) | Relative sensitivity (I) |
|---|---|---|---|
| Ticarcillin(tic) | 73 (40.6%) | 104 (57.8%) | 3 (1.7%) |
| Piperacillin (pip) | 31 (17.2%) | 118 (65.6%) | 31 (17.2%) |
| Carbenicillin (cb) | 77 (42.8%) | 95 (52.8%) | 8 (4.4%) |
| Meropenem (me) | 22 (12.2%) | 147 (81.8%) | 11 (6.1%) |
| Tetracycline (te) | 94 (52.2%) | 84 (46.7%) | 2 (1.1%) |
| Erythromycin(e) | 95 (52.8%) | 64 (35.6%) | 21 (11.7%) |
| Chloramphenicol (c) | 26 (14.4%) | 140 (77.8%) | 14 (7.8%) |
| Amikacin (am) | 14 (7.8%) | 161 (89.4%) | 5 (2.8%) |
| Tobramycin (tob) | 19 (10.6%) | 154 (85.6%) | 7 (0.6%) |
| Novobiocin (nb) | 123 (68.3%) | 56 (31.1%) | 1 (1.7%) |
| Nalidixic acid (na) | 93 (51.7%) | 84 (46.7%) | 3 (1.7%) |
| Rifampin (ra) | 124 (68.9%) | 56 (31.1%) | 0 (0.0%) |
| Ceftazidime (caz) | 70 (38.9%) | 98 (54.4%) | 12 (6.7%) |
| Cefotaxime (ctx) | 76 (42.2%) | 94 (52.2%) | 10 (5.6%) |
| Cefepime (fep) | 55 (30.6%) | 119 (66.1%) | 6 (3.3%) |
| Cefuroxime (xm) | 86 (47.8%) | 65 (36.1%) | 29 (16.1%) |
| Ciprofloxacin (cp) | 56 (31.1%) | 107 (59.4%) | 17 (9.4%) |
| Norfloxacin (nor) | 58 (32.2%) | 114 (63.3%) | 8 (4.4%) |
Table 3. Results of antibiogram tests and the isolates resistance.
110 (61.1 %) of isolates were simultaneously resistant to at least three different antibiotics and 70 (38.9%) of them were resistant to less than two antibiotics. According to the results no samples were resistant to all antibiotics. The results of antibiogram tests and presence of acrA gene investigation showed that the highest antibiotic resistances among the strains with the acrA gene were against piperacillin and rifampin (64.1%), novobiocin (63.0%), tetracycline (48.9%), nalidixic acid and erythromycin (46.7%), cefuroxime (42.4%), carbenicillin (40.2%), cefotaxime (39.1%), ceftazidime (38.4%), ticarcillin (35.9%), norfloxacin (31.5%), Ciprofloxacin (30.4%), Cefepime (29.3%), tobramycin (12.0%), chloramphenicol (13.0%), meropenem (7.3%) and amikacin (6.5%) respectively.
Likewise these results indicated that among the strains with the acrB gene the highest antibiotic resistance were against novobiocin and rifampin (66.6%), tetracycline (52.6%), nalidixic acid and erythromycin (51.9%), cefuroxime (43.0%), carbenicillin (41.5%), cefotaxime (40.7 %), ticarcillin and ceftazidime (37.8%), norfloxacin (33.3%), cefepime (31.1%), ciprofloxacin (30.4%), chloramphenicol (15.6%), piperacillin (14.8%), meropenem (13.3 %), tobramycin (12.6%) and amikacin (9/6%). According to the results of antibiogram tests and presence of tolC gene, the isolates with the tolC gene had the most antibiotic resistances to rifampin (69.6%), novobiocin (68.8%), tetracycline (54.4%), erythromycin (53.6%), nalidixic acid (52.0%), cefuroxime (47.2%), cefotaxime (40.8%), carbenicillin (40.0%), cefotaxime (39.2%), ticarcillin (38.4%), norfloxacin (32.8%), ciprofloxacin (31.2%), cefepime (30.4%), chloramphenicol (16.0%), piperacillin (15.5%), meropenem (14.4%), tobramycin (10.4%) and amikacin (7.2%).
Comparison of antibiogram tests results with One-way and two-way ANOVA tests to assess the significance of the relationship between antibiotic resistance and the presence of efflux pump genes revealed that there was a significant correlation (p<0.05) between the presence of acrA gene and resistance to antibiotics (except carbenicillin and meropenem). There was a significant correlation between the acrB gene and resistance to all antibiotics except carbenicillin (p<0.05). Moreover there was a significant correlation between the tolC gene and resistance to all antibiotics except chloramphenicol and cefotaxime (p<0.05). The results indicate isolates had the most resistance to rifampin and novobiocin. These antibiotics are transferred by the other path apart from efflux pump, so this pump is ineffective in promoting their resistance. The results indicate there was a significant correlation (p<0.05) between the presence of efflux pump and resistance to used antibiotics but there were no correlation between the presence of this pump and carbenicillin, meropenem, chloramphenicol and ramphenicol, cefotaxime, rifampin and novobiocin (p>0.05).
Discussion
Antibiotic resistance in E. coli has been reported worldwide and increases in resistance rates in this bacterium have raised concerns in both developing and developed countries. Therefore in order to avoid treatment failure and control the infections antibiogram testing should be performed prior to prescribing antibiotics and the indiscriminate use of antibiotics should be avoided. According to results of antibiotic resistance pattern in this study, it was found that amikacin (89.4%), tobramycin (85.6%), meropenem (81.7%), chloramphenicol (77.8%) and in the next levels piperacillin (65.6%), cefepime (66.1%), norfloxacin (63.3%) remain effective at treating E coli. High resistance of Gram-negative bacteria to most examined antibiotics in this study was compatible with the findings of the previous study and it can be considered as a warning alarm for increase in commonly prescribed antibiotics resistance in clinical practice. Various studies in different parts of Iran indicate high resistance against different antibiotics in E. coli isolated from patients. Farshad et al. investigated antibiotic resistance pattern in E. coli isolated from urinary tract infections and found that the resistance of samples to tetracycline, gentamicin, ciprofloxacin and amikacin were 70.8 %, 15.6 %, 8.3% and 3.0% respectively [14].
In the current study, multidrug -resistance (MDR) was observed in more than 80 percent of samples. MDR define as resistance to at least one antibiotic out of three or more antimicrobial categories used for treatment [15]. Widespread misuse of various antibiotics imposes direct pressure on the emergence of resistant strains. Several studies have documented presence of MDR E. coli. Anvarinejad et al. performed a study on 90 E. coli strains isolated from the children aged from 1 month to 14 years with urinary tract infection. Results showed 77% of the isolates were resistant to three or more antibiotics. The predominant pattern among these strains (14.4%) included resistance to ampicillin, cotrimoxazole and tetracycline which repeated among 13 strains [16]. Wagner et al. characterized MDR E. coli isolates from urinary tract infections in dogs [17]. Rzewuska et al. determined the antimicrobial susceptibility of E. coli isolates associated with various types of infections in dogs and cats. According to their results the frequency of MDR E. coli isolation (66.8% of isolates) is alarming [18]. These results similar to results of the present study show MDR E. coli are becoming a global public health concern. These findings raise a warning about the increased prevalence of antibiotic resistance. Design of new drugs requires better identification and understanding the intrinsic mechanisms of resistance such as efflux pumps. Therefor in this study genes acrA, acrB and tolC were selected for investigation because of their importance in the incidence of antibiotic-resistance. In this study, 51.1%, 75.0% and 69.4% of isolates had genes acrA, acrB and tolC respectively. In a study conducted by Swick et al. they measured acrA, acrB, tolC, mdfA, and norE expression in E. coli clinical isolates by using real-time PCR. Their findings suggest acrAB overexpression is an indicator of multidrug resistance [19]. Tikhonova and Zgurskaya analyzed interactions between the inner and outer membrane components of the tri-partite multidrug efflux pump AcrAB-TolC from E. coli. Results showed that antibiotics, the substrates of AcrAB-TolC, stabilize interactions within the complex [20].
There was a significant correlation between the presence of efflux pumps and resistance to used antibiotics in this study (except carbenicillin, meropenem, chloramphenicol, cefotaxime, rifampin and novobiocin). Among the used antibiotics in this research, resistance to meropenem, and chloramphenicol was minimum (with 12.2% and 14.4% resistance) that maybe because efflux pumps have no role in the development of resistance to these antibiotics. Strains had the mid-resistance to carbenicillin and cefotaxime (42.8% and 42.2% resistance respectively). According to the results which indicate disaffiliation between the presence of resistance to this antibiotic and efflux pump, it can be concluded that this amount of resistance to these antibiotics is due to other ways of resistance development.
During comparing the results of different percentage of antibiogram test with similar experiments, it should be noted that regional differences in different parts of the world, or even a country, provides different therapeutic response to antimicrobial drugs. The origin of these differences can be attributed to genetic variation between humans or strains in various regions. So regular and continuing studies should be conducted worldwide. According to the obtained results, it can be conclude that several factors such as frequent and irrational use of antibiotics, enzymatic mutations as well as transfer resistance through plasmids decreased effectiveness of antibiotics in treating common infections. The results suggesting that increasing rate of resistance to antibiotics have become a major challenge for treating patients and widespread occurrence of highly resistant E. coli in developing countries, including Iran, in recent years, has become a major concern for public health and human societies.
References
- Odonkor ST, Addo KK. Bacteria resistance to antibiotics: recent trends and challenges. Int J Biol Med Res 2011;2: 1204 -1210.
- Hulscher ME, Grol RP, van der Meer JW. Antibiotic prescribing in hospitals: a social and behavioural scientific approach. Lancet Infect Dis 2010; 10: 167-175.
- Schippa S, Conte MP. Dysbiotic events in gut microbiota: impact on human health. Nutrients 2014;6:5786-5805.
- Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol 2010; 107: 243-274.
- Moreira MAS, Odrigues PCF, Tomaz RS, Moraes CA. Multidrug efflux systems in Escherchia coli and enterobacter cloacae obtained from wholesome broiler carcasses. Braz J Microbiol 2009;40:241-247
- Rosner JL, Martin RG. Reduction of cellular Stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J Bacteriol 2013;195:1042-1050.
- Liu JH, Pan YS, Yuan V, Wu H, Hu GZ, Chen YX. Genetic variations in the active efflux pump genes acrA/B and tolC in different drug-induced strains of Escherichia coli CVCC 1547. Genet Mol Res 2013;12: 2829-2836.
- Nikaido H, Zgurskaya H. AcrAB and related multidrug efflux pumps of Escherichia coli. J Mol Microbiol Biotechnol 2001;3: 215-218.
- Boerlin P, Travis R, Gyles CL, Smith RR, Lim NJH, Nicholson V, McEwen SA, Friendship R, Archambault M. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol 2005;71:6753-6761.
- Schroeder CM, Zhao C, DebRoy C, Torcolini J, Zhao S, White DG, Wagner DD, McDermott PF, Walker RD, Meng J. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl Environ Microbiol 2002;68: 576–581.
- Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, McDermott PF. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg Infect Dis 2012; 18: 741-749.
- Hojati Z, Salehi Z, Motovali-Bashi M, Korbekandi H, Jami S. Molecular analysis of the clavulanic acid regulatory gene isolated from an Iranian strain of Streptomyces clavuligerus, PTCC 1709. Cell J 2011;13:179-186.
- Plichart C, Sechan Y, Davies N, Legrand AM. PCR and dissection as tools to monitor filarial infection of Aedes polynesiensis mosquitoes in French Polynesia. Filaria J 2006;5:1-9.
- Farshad S, Ranjbar R, Anvarinejad M, Shahidi M, Hosseini M. Emergence of multidrug resistant strains of Eschetichia coli isolated from urinary tract infection. Open Conf Proc J 2010;1:192-196
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268-281.
- Anvarinejad M, Farshad Sh, Emamghoreishi F, Hoseini M. Investigating the frequency of multi-drug resistant strains of Escherichia coli isolated from urinary tract infection in children. Pars Journal of Medical Sciences 2012;9:17-23.
- Wagner S, Gally DL, Argyle SA. Multidrug-resistant Escherichia coli from canine urinary tract infections tend to have commensal phylotypes, lower prevalence of virulence determinants and ampC-replicons. Vet Microbiol 2014;169:171-178.
- Rzewuska M, Czopowicz M, Kizerwetter-Swida M, Chrobak D, Błaszczak B, Binek M. Multidrug resistance in Escherichia coli strains isolated from infections in dogs and cats in Poland (2007–2013). SCI World J 2015; 2015:1-8.
- Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother 2011; 55: 921-924.
- Tikhonova EB1, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem 2004; 279: 32116-32124.