ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 20
Investigation of the subcellular localization of HPeV 3A and 2A proteins
Arsalan Salimi* and Siamak Amirfakhri
School of Biological Science, University of Essex, Colchester, UK
- *Corresponding Author:
- Arsalan Salimi
School of Biological Science
University of Essex Colchester,UK
Accepted date: August 03, 2017
Background: Our study investigated the sub-cellular localization of the Parechovirus HPeV 3A nonstructural protein.
Methods: Primers were designed for making wild type HPeV1 and CAV9 2A, 2B, 2C, 2BC and 3A proteins to use for PCR and the pGEM-T Easy vector. The QuikChange II site-directed mutagenesis kit was used to mutate pEGFP-3A using primers. Cells were grown on cover slips and visualized using an Olympus BX41 fluorescence microscope.
Results: We found that EGFP-3A co-localized partially with Golgi and ERGIC. The mutagenesis analysis suggests that the amino acids 39 to 86 of 3A affect distribution and are essential for localization.
Conclusions: Interestingly, co-expression of HPeV 2A with HPeV 3A caused 2A translocation to the nucleolus. The deletion of the TM region in EGFP-2A prevents re-localization.
Keywords
Subcellular localization, HPeV 3A, HPeV 2A
Introduction
Parechoviruses belong to the Picornavirus family. There are currently 16 serotypes of human Parechoviruses (HPeVs) which are numbered from 1 to 16 [1,2]. Among the 16 types of HPeV, HPeV1 is the most common type seen worldwide and mainly effects young children [3].
The genome of Picornaviruses is monocistronic, meaning that it encodes a single polypeptide. Viral proteases cleave the single polypeptide to produce precursor proteins P1-P3. These precursor proteins are in turn cleaved to form the nonstructural and structural proteins. The non-structural proteins include the followings: 2A, 2B, 2C, 2BC, 3A, 3AB, 3C and 3D. Structural proteins include the virion proteins (VPI-VP4). P1 contains the VP1-4 proteins, P2 the 2A-2C proteins, and P3 the 3A-3D proteins [4].
The 3A non-structural protein and its 3AB precursor are both derived from P3. Picornaviruses 3A proteins are poorly conserved among Picornaviruses. However, 3A proteins are often associated with inhibition of ER-to-Golgi transport helping the viruses to escape from the immune system of the host by inhibiting MHC class I [5-7]. Enteroviruses 3A proteins are involved in reducing the production of betainterferon, IL-8, and IL-6 in host cells [8]. In some viruses, such as Avian Encephalomyelitis Virus (AEV), 3A enhances the permeability of the cell membrane and hampers cellular secretion [9]. In other Picornaviruses, such as FMDV, cellular secretion remains uninhibited by the 3A proteins. According to Wessels et al. Enterovirus 3A proteins block protein trafficking by inhibiting anterograde transport [10,11]. Individual expression of the Enterovirus 3A protein also leads to the swelling of the ER [12].
In HRV14, HAV, Enterovirus 71 and Theiler’s virus, 3A proteins fail to block the secretary pathway [13]. In PV infected cells, 3A protein blocks protein transport from ER to the Golgi complex by swelling the ER cisternae [14]. Immunofluorescence studies show that the 3A protein of FMDV also binds to the ER, but it does not block the transport of TsO45 GYFP protein to the cell surface [9]. An analysis of several 3A proteins from 15 genera showed that most bind to the protein ACBD3 to recruit PI4KIIIβ to replication complexes [15].
The 2A region is another diverse part of the polyprotein and quite different types of 2A protein are seen in different Picornaviruses. Some 2A proteins in Picornaviruses are involved in the separation of the P1 structural proteins from the P2 and P3 non-structural proteins, but two different mechanisms have been recognized. In other Picornaviruses 2A proteins do not seem to have this function [16,17]. Three 2A types occur frequently in Picornaviruses, 2Apro (a chymotrypsin-like protease found in Enterovirus and Sapelovirus), proteins with an NPGP motif, and Hbox/NC proteins [18].
2Apro is a chymotrypsin-like cysteine protease and causes cleavage at its N terminus, between VP1 and 2A [19]. In addition, it also cleaves several cellular factors, including eukaryotic initiation factor-4G, TATA-binding protein [ 12,20,21]. This leads to shut off the host cell protein synthesis which will allow the virus to use the resources of the cell and may also overcome anti-virus responses in the cell. Long term expression of either 2Apro or 3Cpro induces caspase activation and thus eventually death by apoptosis [20]. PV 2Apro also has a role in the alteration of proteins and RNA trafficking between the nucleus and cytoplasm [22]. Expression of PV 2Apro in uninfected cells causes the cytoplasmic re-localization of some host nuclear factors, initially suggesting that it may be responsible for the degradation of the nuclear pore complex proteins (Nups) during infection [23]. Little work has been done on the effect of Picornavirus infection on the cytoskeleton. PV has been reported to associate and rearrange with the cytoskeleton and intermediate filaments. It was reported that disruption of the cytoskeletal network may be due to the cleavage of certain cellular proteins by the 3C proteinase 3Cpro of Poliovirus [24].
Another picornaviral protease, Rhinovirus 2Apro, has been implicated in the destruction of cytokeratin 8 in virus-infected HeLa cells [25]. Members of the Cardiovirus genus have an NPGP 2A, but this seems to be fused to a different type of protein [26]. The Hbox/NC type of 2A is related to a family of mammalian proteins involved in cell proliferation. It doesn’t seem to be involved in polyprotein processing [27]. Little is known about the Picornavirus 2A Hbox/NC proteins. Samuilova et al. found that the HPeV1 2A showed a dispersed cytoplasmic staining pattern with some perinuclear concentration in most infected cells. However, a diffuse nuclear staining pattern was also observed in some infected cells at a late stage of infection. Viral RNA was partly colocalized with 2A in the perinuclear region. Using North- Western blotting, gel retardation and UV cross linking techniques, it was observed that 2A possesses RNA binding activity and recognizes the 3´ untranslated region (UTR) of the viral genome with highest affinity [28].
PV 2Apro also has a role in alteration of protein and RNA trafficking between the nucleus and cytoplasm [22]. Expression of PV 2Apro in uninfected cells causes the cytoplasmic re-localization of some host nuclear factors, initially suggesting it may be responsible for the degradation of the nuclear pore complex proteins (Nups) during infection which was later confirmed [22,23]. The multifunctional 2A and 3C proteins are involved in the breakdown process of the cytoskeleton at the time of cell lysis. Many picornaviruses have an NPGP (Asparagine-proline-glycine-proline) motif at the 2A/2B boundary and this is responsible for separating the capsid and non-structural regions during translation [29]. This is not due to proteolysis, but is a translational effect due to ribosome skipping, so that no peptide bond is formed between NPG and the following P, but then translation continues [ 29,30].
The aim of this study was to investigate the role of these proteins in the HPeV-infected cell, mainly using EGFP/ mCherry fusions.
Materials and Methods
Primer design for HPeV1 and CAV9, 2A, 2B, 2C, 2BC and 3A proteins
Primers were designed for making wild type HPeV1 and CAV9 2A, 2B, 2C, 2BC and 3A proteins to use for PCR and the pGEM-T Easy vector. Primers also were designed to introduce mutations into specific regions of the HPeV and CAV9 2B, 2C, 2BC and 3A proteins using PCR and the pGEM-T Easy vector. Pairs of overlapping oligonucleotides were also designed to insert specific motifs at the C-terminus of EGFP. Restriction enzyme cutting sites were introduced into primers to clone PCR fragments.
Mutation of pEGFP-3A
The QuikChange II site-directed mutagenesis kit was used to mutate pEGFP-3A using primers. Mutagenesis reactions were prepared in thin-walled PCR tubes on ice: 5.0 μL of 10x reaction buffer, 1 μL DNA (ng amount variable), 2.5 μL of mutagenic primer #1 (at 50 ng/μL), 2.5 μL of mutagenic primer #2 (at 50 ng/μL), 1.0 μL of dNTP mix, PCR-quality water to a final volume of 50 μL were mixed then 1.0 μL PfuTurbo DNA polymerase (at 2.5 U/μL) was fused. Tubes were placed in the cycler to begin the PCR reaction.
PCR, colony PCR
To make wild type and mutations constructs, PCR was performed in a 50 μL volume, thin-walled 200 μL PCR tube. The reaction mixture consisted of 5 μL 10x PCR buffer including Mg2+, 1 μL 10 mM dNTPs, 1 μL DNA (100 ng), 1 μL of each primer (50 ng), 1 μL (2.5 units) pfu polymerase and 40 μL of sterile distilled water.
Colony PCR is designed to quickly screen for plasmid inserts directly from E. coli colonies and can be used to determine insert size and/or orientation in the vector. The colony PCR procedure involved mixing 38 μL sterile distilled water, 5 μL 10x PCR buffer, 3 μL 25 mM MgCl2, 1 μL 10 mM dNTPs, 1 μL (50 ng) forward primer, 1 μL (50 ng) reverse primer and 0.2-1 μL Taq polymerase to give 50 μL total volume, then a single colony was fused directly to the mixture.
Ligation into pGEMT-easy vector
Ligation reactions were done in a 10 μL volume. 2 μL of the purified PCR product, 1 μL of pGEMT-Easy vector, 5 μL of 2x rapid ligation buffer, 1 μL of T4 DNA ligase and 1 μL of distilled water were mixed. The mixture was then left at room temperature for 1 h.
The clone pEGFP-C1 was cut by double digestion (e.g. with HindIII and BamHI) and purified ligation into pEGFP-C1/PmCherry-C1 vector
Ligations were performed using T4 DNA ligase in a 20 μL reaction containing a 3:1 ratio of insert to vector. 2 μL of the vector DNA, 4 μL of insert DNA, 2 μL of 2x rapid ligation buffer, 1 μL of T4 DNA ligase and 11 μL of distilled water were mixed.
Cell splitting
GMK cells were propagated in 25 cm2 tissue culture flasks containing 5 ml MEM (Minimal Eagles Medium) with 10% foetal calf serum and 100 mg/L gentamycin. For splitting, the medium was renewed and the cells washed twice with PBS. 1 mL Tryple E was fused and incubated with rocking for 5 min to detach the cells. 15 mL medium was then fused and 35 mL portions of the solution divided into 3 new flasks. The flasks were incubated at 37°C for 3-7 d.
Transfection
Cells were grown on cover slips in 6 well plates for 24 h. The cells were washed with serum-free medium. 5 μL DNA and 95 μL serum free medium were fused to a tube and incubated for 30 min. In a second tube, 5 μL Lipofectin TM and serum free medium were combined. The tubes were mixed and incubated for 30 min. 1 mL serum-free medium was fused. The liquid was applied to one cover slip which was incubated overnight at 37°C. The cover slip was washed with PBS and mounted on a microscope slide in 100 μL PBS.
Fluorescent microscopy
Cells were visualized using an Olympus BX41 fluorescence microscope using 60 x filters. The magnification was set at 60,000 x with a snap shot time of 100 ms-1 s for GFP and 5000 ns-10 ms for Hoechst.
Results
Intracellular localization of the 3A protein from HPeV1 and CAV9
To investigate the localization of 3A HPeV1 protein and compare with 3A of a distantly related Picornavirus the Enterovirus CAV9, fusion constructs were produced. 3A was tagged to the C-terminus of EGFP or m-Cherry.
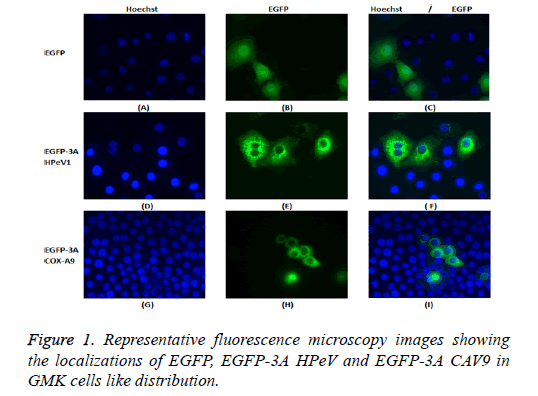
GMK cells were transfected with the constructs, EGFP-3A HPeV1 and EGFP-3A CAV9. Constructs were transfected using LipofectinTM and nuclei were stained 2 d after transfection with Hoechst stain (33342). Fluorescence microscopy results are shown in Figure 1.
It can be seen that EGFP localizes throughout the cell in most cells, although there is a concentration in the nucleus in some cells. In contrast, EGFP-3A HPeV and EGFP-3A CAV9 are largely excluded from the nucleus and seem to be present in perinuclear structures.
Analysis of HPeV1 3A regions involved in localization
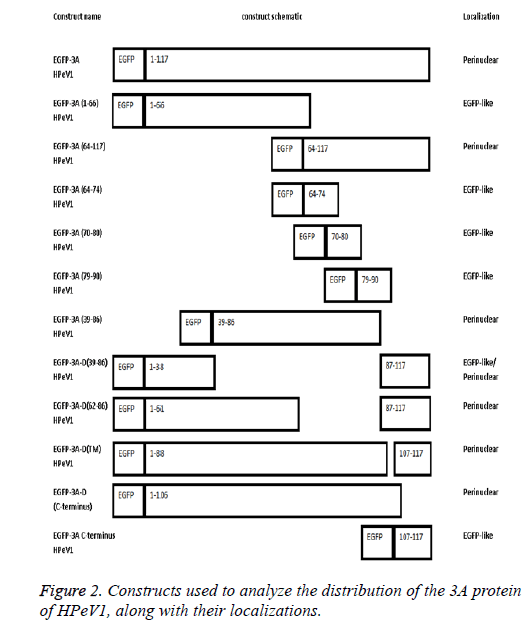
The length of 3A HPeV is 117 amino acids long. To investigate the basis of localization further, we produced several EGFP-3A constructs (Figure 2). The C-terminus of the HPeV1 3A protein is rich in acidic and basic amino acids in both HPeV and Ljungan Virus (LV). To find whether these contribute to localization, the C-terminal 11 amino acids of the HPeV 3A protein (KIFKKEESKDE) and C-terminal amino acids of the LV protein (KKEKKEEERQE) were fused to the C-terminus of EGFP. A version of EGFP-3A lacking the final 11 amino acids was also produced. Transfection results showed that the C-terminal fusions had the same localization as EGFP itself. In addition, the construct lacking the C-terminal had the same distribution as EGFP-3A. This suggests that the C-terminus of HPeV, and probably LV, 3A doesn’t affect distribution.
To find whether the 3A N-terminus (1-66) contribute to localization, we fused amino acids 1 and 66 to EGFP. Transfection results showed that the EGFP construct with these amino acids region had the same distribution as EGFP. This suggests that amino acids 1 to 66 are not sufficient to give 3Alike localization. In contract amino acids 64-117 fused to EGFP give 3A.
To find whether the 3A transmembrane domain (amino acids) contributes to localization, we made an EGFP construct lacking this domain (amino acids 89-106). Surprisingly, transfection results showed that the produced EGFP protein had a very similar distribution to EGFP-3A although some fluorescence is more dispersed. This suggests that the transmembrane domain of HPeV doesn’t fully define the distribution. We also made constructs to understand the role of amino acids 39 to 86 in localization. We made two EGFP constructs; one contained only amino acids 39 to 86 from 3A and the other contained 3A without amino acids 39 to 86. Transfection results showed that only amino acids 39 to 86 gave the same distribution as EGFP-3A, while the construct lacking the region from 39 to 86 had a more dispersed localization, but with some perinuclear concentration. This suggests that the region from 39 to 86 of HPeV affect distribution and is important for localization. As amino acids 64-117 seem to be important, the results suggest that the 64-86 region will give the same distribution as EGFP-3A.
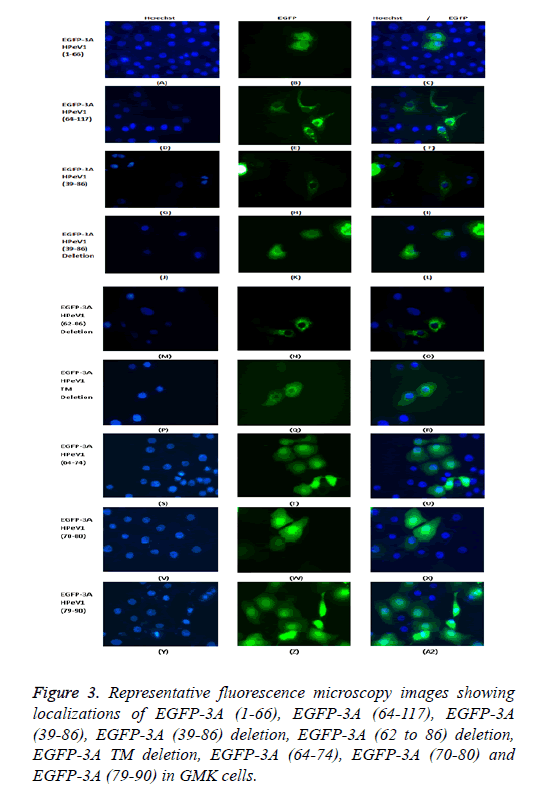
To find which amino acid or group of amino acids in the region from 64 to 86, give 3A localization, we divided this region into 3 overlapping parts and fused each sequence to the C-terminus of EGFP. Each part contains 11 amino acids SREVVDWFSDK, WFSDKIKSMLS and LSFVERNKAWLT. Transfection results showed that all the constructs had the same localization as EGFP itself. This suggests that the 11 amino acid sequences (SREVVDWFSDK), (WFSDKIKSMLS) and (LSFVERNKAWLT) cannot work individually to give the correct 3A localization. A version of EGFP-3A lacking the region from 62 to 86 was also produced. Transfection results showed that the construct lacking the region from 62 to 86 had the same distribution as EGFP-3A. This suggests that the region from 64 to 86 of HPeV 3A doesn’t infect affect distribution. The results suggest that the amino acids 39-61 and the transmembrane region, both contribute to localization.
To confirm that the above constructs have same distribution to EGFP-3A HPeV1, we co-transfected with m-Cherry-3A HPeV1. Apart from the transmembrane domain construct, all the EGFP constructs had a similar distribution to mCherry-3A. This may be due to the ability of 3A to form dimmers which could allow the mutants proteins to be relocated by dimerization with m-Cherry-3A.
To investigate the basis of localization further, we produced several EGFP-3A constructs. The C-terminus of the HPeV1 3A protein is rich in acidic and basic amino acids in both HPeV and LV. To find whether these contribute to localization, the Cterminal 11 amino acids of 3A (KIFKKEESKDE) and Cterminal amino acids of the LV protein (KKEKKEEERQE) were fused to the C-terminus of EGFP. A version of EGFP-3A lacking the final 11 amino acids was also produced. Transfection results showed that the C-terminal fusions had the same localization as EGFP itself. In addition, the construct lacking the C-terminus of 3A had the same distribution as EGFP-3A. This suggests that the C-terminus of HPeV, and probably LV, 3A doesn’t affect distribution.
To find whether the 3A N-terminus (1-66) contributed to localization, we fused amino acids 1 and 66 to EGFP. Transfection results showed that the EGFP construct with these amino acids region had the same distribution as EGFP. This suggests that amino acids 1 to 66 aren’t sufficient to give 3Alike localization. In contrast, amino acids 64-117 fused to EGFP give 3A-like distribution.
To find whether the 3A transmembrane domain (amino acids) contributes to localization, two further constructs were assembled an EGFP construct lacking this domain (amino acids 89-106). Surprisingly transfection results showed that the EGFP protein produced had a very similar distribution to EGFP-3A although some fluorescence is more dispersed. This suggests that the transmembrane domain of HPeV does not fully define the distribution. Two further constructs were assembled to understand the role of amino acids 39 to 86 in localization. Two further EGFP constructs were assembled; one contained only amino acids 39 to 86 from 3A and the other contained 3A without amino acids 39 to 86. Transfection results showed that only amino acids 39 to 86 gave the same distribution as EGFP-3A, while the construct lacking the region from 39 to 86 had a more dispersed localization, but with some perinuclear concentration. This suggests that the region from 39 to 86 of HPeV affect distribution and is important for localization. As amino acids 64-117 seem to be important, the results suggest that the 64-86 region will give the same distribution as EGFP-3A.
To find which amino acid or group of amino acids in the region from 64 to 86 affect 3A localization, we divided this region into 3 overlapping parts and fused each sequence to the Cterminus of EGFP. Each part contains 11 amino acids SREVVDWFSDK, WFSDKIKSMLS and LSFVERNKAWLT. Transfection results showed that all the constructs had the same localization as EGFP itself. This suggests that the 11 amino acid sequences (SREVVDWFSDK), (WFSDKIKSMLS) and (LSFVERNKAWLT) cannot work individually to give the correct 3A localization. A version of EGFP-3A lacking the region from 62 to 86 was also produced. Transfection results showed that the construct lacking the region from 62 to 86 had the same distribution as EGFP-3A (Figure 3). This suggests that the region from 64 to 86 of HPeV 3A doesn’t in fact affect distribution. The results seem to suggest that the amino acids 39-61 and the transmembrane region both contribute to localization.
To confirm that the above constructs have same distribution to EGFP-3A HPeV1, we co-transfected with m-Cherry-3A HPeV1. Apart from the transmembrane domain construct, all the EGFP constructs had a similar distribution to mCherry-3A. This may be due to the ability of 3A to form dimers which could allow the mutants proteins to be relocated by dimerization with m-Cherry-3A.
Co-transfection of EGFP-3A or m-Cherry-3A protein with m-Cherry-2A, m-Cherry-2B and m-Cherry-2C HPeV1 non-structural proteins
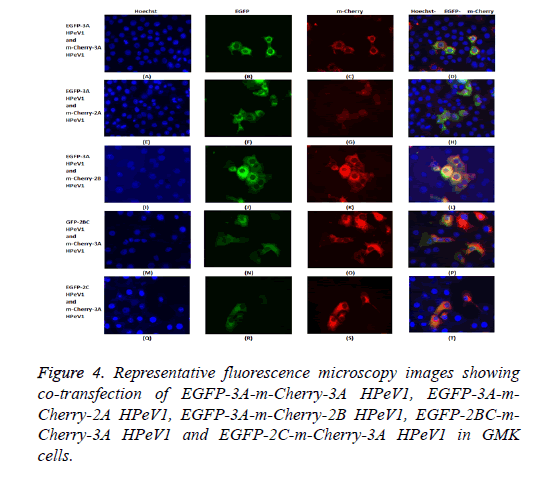
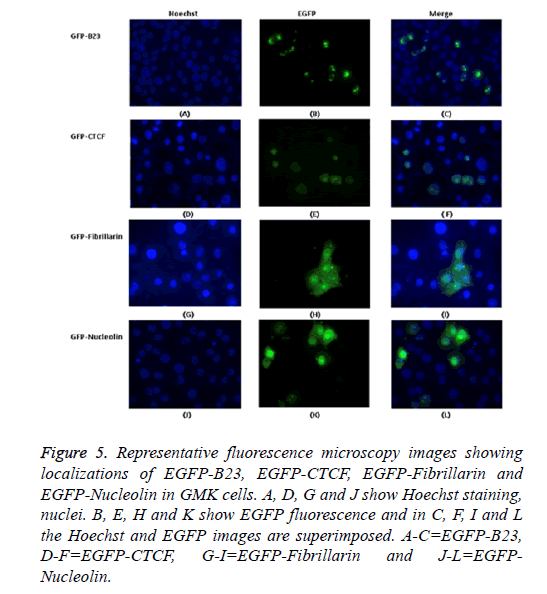
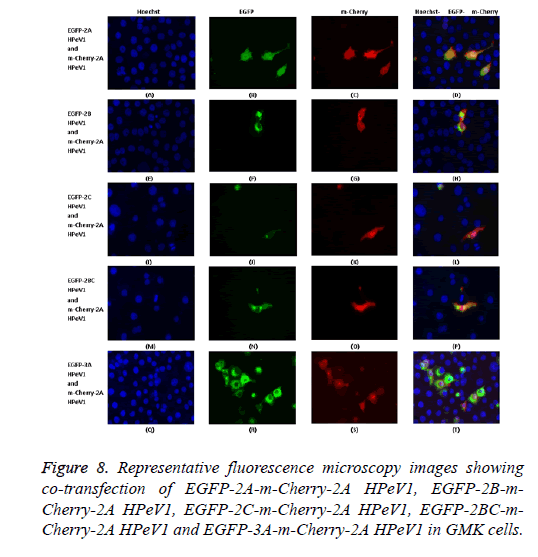
The possible co-localization of 3A and other non-structural proteins was investigated using EGFP-3A/m-Cherry-3A and m-Cherry-2A, m-Cherry-2B, EGFP-2BC and EGFP-2C ( Figure 3). There was some co-localization of 3A with all these proteins except 2A. Interestingly, the presence of EGFP-3A changed the localization of 2A and some of this protein localized in regions of the nuclei, presumably corresponding to nucleoli (Figure 4). To investigate co-localization of 2A with other non-structural HPeV proteins the m-Cherry-2A construct was co-transfected with EGFP-3A, EGFP-2B, EGFP-2C and EGFP-2BC. The other non-structural proteins had a more punctuate distribution than EGFP-2A and these proteins were excluded from the nucleus, which EGFP-2A is seen in the nucleus. There was no co-localization between 2A and the other non-structural proteins. However, the same redistribution of m-Cherry-2A was seen when EGFP-3A was co-transfected ( Figure 4). Intense spots were observed in the nucleus, which seemed to correspond to the nucleoli. Co-transfection of EGFP-3A HPeV1 and m-Cherry-2A showed nucleolus signal dots within the nucleus. For understanding better the function of these nucleolus dots we investigated the structure of EGFPB23, EGFP-CTCF, EGFP-Fibrillarin and EGFP-Nucleolin (as nucleolar proteins) in cells that were transfected with m- Cherry-3A and m-Cherry-2A HPeV1 to identifying the basis of changes in structure of nuclear proteins or m-Cherry-3A/ Cherry-2A HPeV1 or investigate which there is any interaction between nuclear proteins and m-Cherry-3A/m-Cherry-2A HPeV1. Results showed that m-Cherry-3A HPeV1 didn’t have any effect on the above nuclear proteins (Figure 5).
Figure 5: Representative fluorescence microscopy images showing localizations of EGFP-B23, EGFP-CTCF, EGFP-Fibrillarin and EGFP-Nucleolin in GMK cells. A, D, G and J show Hoechst staining, nuclei. B, E, H and K show EGFP fluorescence and in C, F, I and L the Hoechst and EGFP images are superimposed. A-C=EGFP-B23, D-F=EGFP-CTCF, G-I=EGFP-Fibrillarin and J-L=EGFPNucleolin.
Intracellular localization of the HPeV1 EGFP and m- Cherry 2A protein
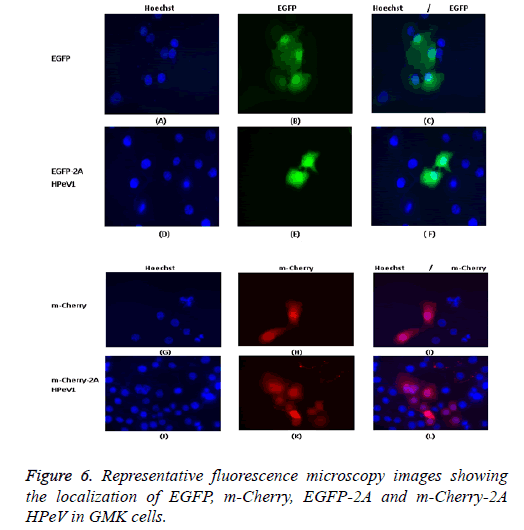
Individually expressed HPeV1 2A is found to be diffused in the cytoplasm and nucleus and its localization doesn’t change upon infection [31]. This is also checked with other kind of methods like electron microscopy. We also confirm these mcherry or EGFP with other proteins and all shows correct distribution so it means m-cherry or EGFP cannot effect on localization of protein itself. So we know m-cherry and EGFP and 2A have all over the cell distribution but if we focus more closely we can see some nuclei dots inside the nuclear when 2A is present in EGFP OR m-cherry. It can be seen that EGFP localizes throughout the cell in most cells although there is a concentration in the nucleus in some cells and EGFP-2A HPeV is also largely located in the cytoplasm. There is a little difference between the EGFP and EGFP-2A localization. Similar results were obtained for m-Cherry and m-Cherry-2A.
In previous studies using EGFP fusions 2A and 3D was found in the cytoplasm and nucleus, whereas 2B, 2C, 2BC, 3A and 3AB were found in the cytoplasm, mainly around the nucleus [ 28,31]. The pattern of transfected 2A-EGFP didn’t change upon infection, and no specific co-localization between the transfected proteins and the viral 2C protein was observed.
One of our aims in this study was to investigate the localization of the 2A HPeV1 to help understand the function of this protein. To understand the localization, we tagged the Cterminus of EGFP or m-Cherry to the 2A protein of HPeV1. GMK cells were transfected with the constructs. Constructs were transfected using Lipofectin TM and nuclei were stained two days after transfection with Hoechst stain (33342). Fluorescence microscopy results are shown in Figure 6. It can be seen that EGFP localizes throughout the cell in most cells although there is a concentration in the nucleus in some cells and EGFP-2A HPeV is also largely located in the cytoplasm. There is little difference between the EGFP and EGFP-2A localization. Similar results were obtained for m-Cherry and m-Cherry-2A.
Analysis of HPeV1 2A regions involved in localization
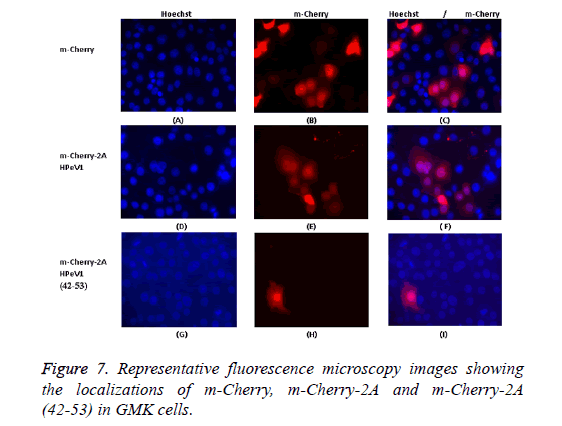
To investigate the basis of localization further, we produced several constructs including EGFP-2A (1-80) (N-terminal 80 amino acids of 2A), EGFP-2A (80-147) (C-terminal 68 amino acids) and EGFP-2A (TM-deletion) to investigate which region contribute to localization. In addition, a 12 amino acids region (ALTGKAKFTKTK) position (42-53) of the HPeV 2A protein, which includes the BXBXXBXB motif, was fused to the Cterminus of m-Cherry. This construct was called m-Cherry-2A (42-53). An m-Cherry-2A construct was also made. Transfection results showed that m-Cherry-2A (42-53) had the same localization as m-Cherry-2A and m-Cherry itself (Figure 7). Transfection results indicated that the EGFP construct with EGFP-2A (1-80), EGFP-2A (80-147) and EGFP-2A (TMdeletion) region had the same distribution as EGFP-2A HPeV1. Therefore the mutations don’t effect distribution.
To confirm that the above constructs have same distribution as EGFP-2A HPeV1, we produced m-Cherry-2A HPeV1. All the above EGFP-2A regions constructs were co-transfected with m-Cherry-2A HPeV. The results showed that all the mutants construct have the same distribution as m-Cherry-2A.
Co-transfection of m-Cherry-2A with EGFP-3A, EGFP-2A, EGFP-2B, EGFP-2BC and EGFP-2C HPeV1 non-structural proteins
To investigate the effect of 2A mutations, the mutant EGFP-2A constructs were co-transfected with m-Cherry-2A. All the mutants co-localized and so the mutations have no effect on localization. To investigate co-localization of 2A with other non-structural HPeV proteins the m-Cherry-2A construct was co-transfected with EGFP-3A, EGFP-2B, EGFP-2C and EGFP-2BC. The other non-structural proteins had a more punctuate distribution than EGFP-2A and these proteins were excluded from the nucleus, which EGFP-2A is seen in the nucleus. There was no co-localization between 2A and the other non-structural proteins. However, the same redistribution of m-Cherry-2A described in chapter 3 was seen when EGFP-3A was co-transfected (Figure 8). Intense spots were observed in the nucleus, which seemed to correspond to the nucleoli.
Regions of 2A and 3A needed for nucleolar localization
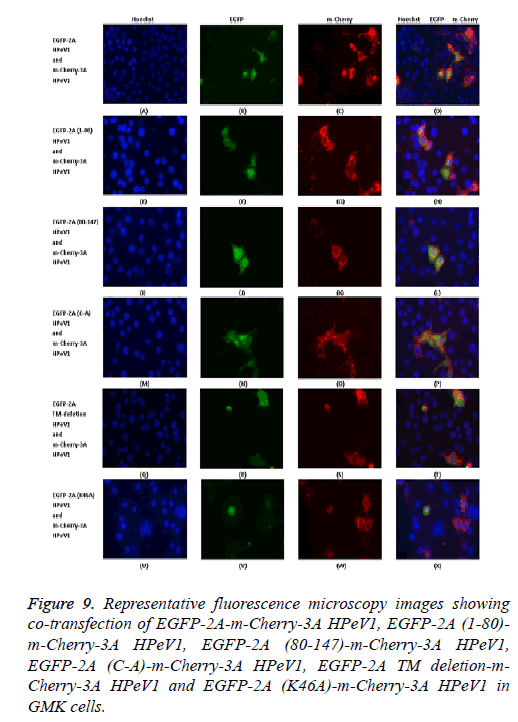
To investigate the relocalization of 2A further, mutant EGFP-2A constructs were co-transfected with m-Cherry-3A ( Figure 9). Both the EGFP-2A (1-80) and EGFP-2A (80-147) construct were not relocalized, suggesting that the relocalization may need more than one region of 2A. The deletion of the TM region prevents relocalization, while the mutation of the cysteine of the NC motif has no effect.
Figure 9: Representative fluorescence microscopy images showing co-transfection of EGFP-2A-m-Cherry-3A HPeV1, EGFP-2A (1-80)- m-Cherry-3A HPeV1, EGFP-2A (80-147)-m-Cherry-3A HPeV1, EGFP-2A (C-A)-m-Cherry-3A HPeV1, EGFP-2A TM deletion-m- Cherry-3A HPeV1 and EGFP-2A (K46A)-m-Cherry-3A HPeV1 in GMK cells.
Analysis of the 3A mutants shown that 3A proteins (1-66) cannot cause relocalization. Relocalization by 64-117 does not seem to be as strong as that caused by the intact protein. Both the 39-86 region construct and the 39-86 deletion could cause some relocalization, suggesting that more than one region can cause relocalization.
m-Cherry or EGFP control in any figures should be similar localization (means all over the cells) and if we are going to add picornavirus non-structural gene in it we expect to have different localization because the region of m-cherry or EGFP have short gene that don’t effect on localization of 3A or 2A. So whatever localization we seen on m-Cherry-3A, m- Cherry-2A or EGFP-3A, EGFP-2A are actual localization of 3A or 2A as a wild type. We were expecting to make so many mutations in region of 3A and 2A to see more relocalizations and changes.
Discussion
The aim of this study was to investigate the subcellular localization of HPeV 3A and 2A proteins and understanding the role of these proteins in the HPeV-infected cell, mainly using EGFP/mCherry fusions. Experiments were performed with GMK cells, because HPeV1, the virus used in this study, is able to replicate in these cells, indicating that the 3A proteins has the ability to fulfil its functions in these cells. Therefore, we characterized the sub-cellular localizations of EGFP-3A and the mutants in these cells.
To study the localization of the 3A protein and investigate which amino acids in 3A are involved in 3A localization several more constructs were made and analyzed. Amino acid position 88 to 106 of 3A is strongly hydrophobic which may indicate possible transmembrane linkage. However, the evidence suggests that EGFP-3A lacking the transmembrane region has a distribution between EGFP and EGFP-3A with significant diffuse staining throughout the cell. We confirmed therefore that the 3A transmembrane region alone cannot give the 3A distribution seen. Therefore, we continued to make several additional constructs to determine the sequences in the HPeV 3A protein which direct it to its cellular location. This seemed to indicate that the region from amino acids 39 to 86 is the critical region for localization of 3A HPeV1, because EGFP-3A without this region is located throughout the cell. In contrast, when only the region from 39 to 86 was fused to the C-terminus of EGFP, this protein was largely excluded from the nucleus and seemed to be present in perinuclear structure, as seen for EGFP-3A. Removing amino acids 62-86 gave a protein with EGFP-3A-like distribution, suggesting that only amino acids 39-61 are important in localization. However, this is not fully consistent with fact that removing positions 1-63 (N-terminus) didn’t seem to greatly affect the localization. The N-terminal deletion construct distribution is similar to EGFP-3A, but there may be some differences. The results may suggest that more than one part of the 3A protein regions is involved in localization. The experiments indicate that amino acids 39-61 and the transmembrane region are important. Amino acids 39-61 include the region that aligns with the helical hairpin in Enterovirus 3A [32]. This region is important in Enterovirus 3A dimerization and if it contributes to HPeV 3A dimerization this could explain why this region is a key to localization. This was also analyzed by co-localization studies. These confirmed that m-Cherry has a similar distribution intermediate to that of EGFP and EGFP-3A. The results supported the idea that the 39-61 regions are important in localization. Possible dimerization of 3A was studied by adding the motif TKL to the C-terminus of m-Cherry-3A. This gives a highly punctuate distribution, presumably due to peroxisomal localization. Surprisingly, this is lost in most of the co-transfection with EGFP constructs which give a periplasmic localization to m-Cherry-3A, possible due to dimerization. However, the EGFP-3A 39-89 deletion has a very different distribution to m-Cherry-3A TKL which adds more support to the importance of this region in dimerization. In the case of Picornaviruses, dimerization is found to be important in non-structural proteins [8,33]. 3A protein in CVB3 (Coxsackievirus B3) form homodimer and this homodimerization in 3A play a key factor for inhibition of protein trafficking and viral RNA [11].
The 2A proteins are diverse among Picornaviruses and don’t have homologous structures and functions for certain nonstructural proteins [34]. 2A proteins of these viruses are also related to certain proteins of cell cycle and cell division such as H-rev107, TIG3 and LRAT which are possibly involved in the control of cell proliferation [18]. The 2A protein of HPeV lacks proteolytic activity and no function has been identified to date, but it is believed that it may have role to play in viral replication [28].
The EGFP-2A has a similar distribution to EGFP itself. Surprisingly, EGFP-2A doesn’t seem to be associated with membranes despite its putative transmembrane domain. The basic motif (BXBXXBXB) doesn’t seem to allow EGFP-2A to localize in the nucleus more than EGFP itself. To further study the localization of the 2A protein, several constructs were made encoding EGFP-2A (1-80), EGFP-2A (80-147), EGFP-2A (TM-deletion) and EGFP-2A (42-53). The evidence suggests that EGFP-2A (1-80), EGFP-2A (80-147), EGFP-2A (TM-deletion) and EGFP-2A (42-53) alone have an almost identical distribution to EGFP-2A. We confirmed this by colocalization of m-Cherry-2A with our constructs. Therefore, none of these regions can change the distribution of EGFP. An m-Cherry-2A was produced for these studies. Our results showed that EGFP-2A and m-Cherry-2A have similar distribution. The m-Cherry-2A distribution is very different from that of EGFP-3A, EGFP-2B, EGFP-2BC and EGFP-2C and there is no co-localization. The most interesting result was obtained with EGFP-3A. We saw dots in the nucleus (apparently in nucleoli) in the m-Cherry-2A and this distribution was only seen when EGFP-3A and m-Cherry-2A when were together. This re-localization doesn’t seem to need the conserved H2 and C90 of 2A, but need the transmembrane domain. The fact that the 2A transmembrane domain doesn’t seem to localize 2A to membranes and is involved in relocalization caused by 3A, may suggest that it is not a transmembrane domain. It could instead be an interaction domain that allows 2A to interact with another protein with nucleolar localization and this takes 2A to the nucleolus.
As the HPeV 2A protein is localized partially to the nucleolus, it is of interest to decipher whether HPeV-2A affects the nucleolus.
The deletion of the EGFP-2A TM region prevents relocalization, while the mutation of the cysteine of the NC motif had no effect. Both the EGFP-3A (39-86 region) construct and the EGFP-3A (39-86 deletion) could cause some relocalization, suggesting that more than one region can cause relocalization. This observation will be followed up by trying to find binding partners of 2A, using proteomics approaches, which may explain the relocalization.
2A proteins of Picornaviruses have varied function as well as varied sizes [18,34]. Different members of the Picornavirus family use 2A for different purposes. One of the functions of 2A is processing of the polyprotein. This involves separation of structural and non-structural proteins of Picornaviruses i.e. P1 from P2 and P3. This activity of 2A is found in Enterovirus and Sapelovirus where it behaves as a trypsin-like cysteine protease [29]. Such 2A proteins with protease activity are notated as 2Apro. Another group of Picornaviruses also use 2A proteins to separate structural and non-structural proteins, not by proteolytic cleavage but by mediating ribosome skipping and thus having an effect on translation. This effect is mediated by an Asn-Pro-Gly-Pro (NPGP) motif [30]. 2A of some Picornaviruses is not associated with polyprotein processing in any manner at all, nor is any similarity seen with other 2A proteins [16,17,31].
The 2A protein of Parechovirus belongs to a group of proteins called H box-NC proteins on the basis of two conserved domains [4]. Several Picornavirus genera have a 2A of this type. These proteins have an Hbox (a motif close to the Nterminus motif comprising of four amino acids that starts with an absolutely conserved H residue followed usually by one large hydrophobic and two small hydrophobic amino acids) and an NC motif (found towards the C-terminus) [4]. Cellular proteins such as H-rev107, Tazarotene-induced gene protein 3 (TIG3) and lecithin retinol acyltransferase (LRAT) also have these motifs [4]. These cellular proteins are possibly involved in control of cell proliferation. The function of Hbox-NC 2A proteins is not yet known. It is known that it binds to the 3’ UTR of viral RNA and though the significance of binding is not known, it is believed that it may have role to play in viral replication [28]. The Kobuvirus 2A can also bind to viral RNA and has a role in replication [35]. Individually expressed HPeV1 2A is found to be diffused in the cytoplasm and nucleus and its localization does not change upon infection [ 31]. 2A of HPeV is also found to be structurally related to certain proteins involved in the cell cycle and cell division such as H-rev107, TIG3 and LRAT [18].
The cellular relatives of 2A have been investigated recently. Two of these are phospholipases and it 2A has a similar function; this may have an effect on ER, ERGIC and Golgi membranes. We also examined which non-structural proteins co-localize with each other. An m-Cherry-2A was produced for these studies. Our results showed that EGFP-2A and m- Cherry-2A have similar distribution. The m-Cherry-2A distribution is very different from that of EGFP-3A, EGFP-2B, EGFP-2BC and EGFP-2C and there is no co-localization. The most interesting result was obtained with EGFP-3A. We saw dots in the nucleus (apparently in nucleoli) in the m-Cherry-2A and this distribution was only seen when EGFP-3A and m- Cherry-2A when were together. This relocalization does not seem to need the conserved H2 and C90 of 2A, but does need the transmembrane domain. The fact that the 2A transmembrane domain does not seem to localize 2A to membranes and is involved in re-localization caused by 3A, may suggest that it is not a transmembrane domain. It could instead be an interaction domain that allows 2A to interact with another protein with nucleolar localization and this takes 2A to the nucleolus.
As the HPeV 2A protein is localized partially to the nucleolus, it is of interest to find whether HPeV-2A affects the nucleolus. The effect of 2A HPeV protein on four nuclear proteins B23, fibrillarin, nucleolin and CTCF was studied. The results showed no effect on fibrillarin, nucleolin or CTCF distribution.
Competing Interests
There is no support or conflict of interest for the present study.
Author’s Contribution
Both authors worked equally for the article.
Acknowledgment
There is no acknowledgment for the present study.
References
- Agol VI, Gmyl AP. Viral security proteins: counteracting host defences. Nat Rev Microbiol 2010; 8: 867-878.
- Barco A, Feduchi E, Carrasco L. A stable HeLa cell line that inducibly expresses poliovirus 2A (pro): effects on cellular and viral gene expression. J Virol 2000; 74: 2383-2392.
- Belov GA, Kovtunovych G, Jackson CL, Ehrenfeld E. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell Microbiol 2010; 12: 1463-1479.
- Benschop KS, Schinkel J, Minnaar RP, Pajkrt D, Spanjerberg L, Kraakman HC, Wolthers KC. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin Infect Dis 2006; 42: 204-210.
- Benschop KS, Williams CH, Wolthers KC, Stanway G, Simmonds P. Widespread recombination within human parechoviruses: analysis of temporal dynamics and constraints. J Gen Virol 2008; 89: 1030-1035.
- Cameron CE, Oh HS, Moustafa IM. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol 2010; 5: 867-881.
- Castello A, Izquierdo JM, Welnowska E, Carrasco L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J Cell Sci 2009; 122: 3799-3809.
- Choe SS, Dodd DA, Kirkegaard K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virol 2005; 37: 18-29.
- Doedens JR, Giddings TH, Kirkegaard K. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol 1997; 71: 9054-9064.
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol 2001; 82: 1013-1025.
- Foeger N, Schmid EM, Skern T. Human rhinovirus 2 2Apro recognition of eukaryotic initiation factor 4GI. Involvement of an exosite. J Biol Chem 2003; 278: 33200-33207.
- Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. J Virol 2012; 86: 3605-3616.
- Hughes PJ, Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J Gen Virol 2000; 81: 201-207.
- Jia XY, Summers DF, Ehrenfeld E. Primary cleavage of the HAV capsid protein precursor in the middle of the proposed 2A coding region. Virol 1993; 193: 515-519.
- Joachims M, Van Breugel PC, Lloyd RE. Cleavage of poly (A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 1999; 73: 718-727.
- Joki-Korpela P, Hyypia T. Parechoviruses, a novel group of human picornaviruses. Ann Med 2001; 33: 466-471.
- Knowles N, Hovi T, Hyypiä T, King A, Lindberg A, Pallansch M, Stanway G. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on taxonomy of viruses. Picornaviridae 2012; 855-880.
- Krogerus C, Samuilova O, Poyry T, Jokitalo E, Hyypia T. Intracellular localization and effects of individually expressed human parechovirus 1 non-structural protein. J Gen Virol 2007; 88: 831-841.
- Liu J, Wei T, Kwang J. Avian encephalomyelitis virus non-structural protein 2C induces apoptosis by activating cytochrome c/caspase-9 pathway. Virol 2004; 318: 169-182.
- Moffat K, Howell G, Knox C, Belsham GJ, Monaghan P, Ryan MD, Wileman T. Effects of foot-and-mouth disease virus non-structural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J Virol 2005; 79: 4382-4395.
- Neznanov N, Kondratova A, Chumakov KM, Angres B, Zhumabayeva B, Agol VI, Gudkov AV. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J Virol 2001; 75: 10409-10420.
- O'Donnell VK, Pacheco JM, Henry TM, Mason PW. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virol 2001; 287: 151-162.
- Ryan MD, Flint M. Virus-encoded proteinases of the picornavirus super-group. J Gen Virol 1997; 78: 699-723.
- Samuilova O, Krogerus C, Poyry T, Hyypia T. Specific interaction between human parechovirus nonstructural 2A protein and viral RNA. J Biol Chem 2004; 279: 37822-37831.
- Sasaki J, Taniguchi K. Aichi virus 2A protein is involved in viral RNA replication. J Virol 2008; 82: 9765-9769.
- Schultheiss T, Emerson SU, Purcell RH, Gauss-Muller V. Polyprotein processing in echovirus 22: a first assessment. Biochem Biophys Res Commun 1995; 217: 1120-1127.
- Seipelt J, Liebig HD, Sommergruber W, Gerner C, Kuechler E. 2A proteinase of human rhinovirus cleaves cytokeratin 8 in infected HeLa cells. J Biol Chem 2000; 275: 20084-20089.
- Strauss DM, Glustrom LW, Wuttke DS. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J Mol Biol 2003; 330: 225-234.
- Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol 2005; 25: 10556-10565.
- Toyoda H, Nicklin MJ, Murray MG, Anderson CW, Dunn JJ, Studier FW, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 1986; 45: 761-770.
- Wessels E, Duijsings D, Lanke KH, van Dooren SH, Jackson CL, Melchers WJ, van Kuppeveld FJ. Effects of picornavirus 3A Proteins on Protein Transport and GBF1-dependent COP-I recruitment. J Virol 2006; 80: 11852-11860.
- Wessels E, Duijsings D, Notebaart RA, Melchers WJ, van Kuppeveld FJ. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-golgi transport. J Virol 2005; 79: 5163-5173.
- Yalamanchili P, Banerjee R, Dasgupta A. Poliovirus-encoded protease 2APro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J Virol 1997; 71: 6881-6886.
- Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, Takeda N, Yamazaki S. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol 1998; 72: 8408-8412.
- Zell R, Seitz S, Henke A, Munder T, Wutzler P. Linkage map of protein-protein interactions of Porcine teschovirus. J Gen Virol 2005; 86: 2763-2768.