ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2022) Volume 33, Issue 1
Kruppel-like factor-4 in macrophages synergistically regulates polarization with glucocorticoids and may ameliorate glucocorticoid resistance.
Hanyu Shi1*, Dawei Yin2*, Luqing Wei3, Liang Sun3
1Hospital of the First Mobile Corps of the Chinese People’s Armed Police Force, Dingzhou, Hebei 073099, China
2Academy of Special Operation of the Ground Force of Chinese People's Liberation Army, Guangzhou, Guangdong 510599, China
3Characteristic Medical Center of the Chinese people’s Armed Police Force, Tianjin 300162, China
- Corresponding Author:
- Liang Sun
Characteristic medical center of the Chinese People’s Armed
Police Force
Tianjin 300162
China
Email: sunliangys@sina.com
Accepted date: December 16, 2021
Keywords
Kruppel-Like Factor-4, Interstitial Lung Disease, Dexamethasone, Macrophage Polarization, Glucocorticoid Receptor, Glucocorticoid Resistance
Introduction
Interstitial Lung Disease (ILD), one of the most common respiratory disorders in elderly people worldwide, is characterized by progressive dyspnea and persistent dry cough [1]. A growing body of evidence suggests that the inflammatory response secondary to repeated injuries and Connective Tissue disease (CTD), such as Scleroderma (SSc), contributes to the development of ILD [2]. Recent work by Hu has shown that lipopolysaccharide (LPS) can induce acute lung injury and pulmonary fibrosis via specific pathways [3,4].
Systemic Glucocorticoids (GCs) are common drugs prescribed to treat ILD, especially CTD-ILD, and mainly work by restricting excessive inflammatory responses and immune overactivation via the Glucocorticoid Receptor (GR) signalling pathways [5]. However, long-term administration contributes to GC resistance [6], such that patients on GCs can develop decreased GC sensitivity and even resistance. Resistance is mainly associated with a constriction of the combination of GR and GCs stemming from various reasons [7,8].
Macrophages involved in the pathogenesis of ILD exhibit diverse heterogeneity. Classically, they display two subsets: the M1 subset (classically activated macrophages) mediating proinflammatory actions and the M2 subset (alternatively activated macrophages) promoting tissue repair and resisting excessive inflammatory responses [9]. Kruppel-Like Factors (KLFs) belong to the zinc finger family of transcription factors, of which KLF4 shares a Cterminal three-zinc-finger DNA-binding domain [10]. KLF4 exerts diverse functions in regulating inflammation and promoting differentiation [10]. Studies conducted by Liao and Saha have shown that up regulated KLF4 could promote macrophage polarization towards the M2 subset [9,11].
While studies have evaluated the cooperative regulation of KLF4 and GR to maintain homeostasis in keratinocytes [12], there has been little evidence about whether KLF4 in macrophages and GCs could synergistically regulate polarization to induce lung tissue repair. In addition, no studies have been found regarding whether KLF4 in macrophages could regulate GR expression concomitant with GCs to ameliorate GC resistance.
In this paper, we subjected ILD macrophages with high and low expression of KLF4 (mKLF4 and shKLF4) to GC to examine whether KLF4 in macrophages synergistically regulates polarization with GCs and ameliorates GC resistance in ILD.
Materials and Methods
Cell culture and reagent
Wild-type murine macrophages (WT RAW264.7) were obtained from Professor Jihong Han of the Academy of Life Sciences, Nankai University, China. RAW264.7 cells were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 12% foetal calf serum (Gibco) and maintained in a conventional incubator (37ºC and 5% CO2). The culture was monitored daily, and subculture was performed when the cell density reached 80-90%. Cells in a good growth state and in the logarithmic growth phase were used in all subsequent experiments. All cells were induced with LPS to construct the ILD model. The final concentrations of LPS and Dexamethasone (DX) as macrophage stimuli were set as 1 μg/mL and 1 μM, respectively, according to previous results [13,14].
KLF4 upregulation and knockdown
The lentiviral vector pLVX-mCMV-ZsGreen-PGK-Puro plasmid (Changsha Youbao Biological Co. Ltd.) was double digested with EcoR I (10 U/μL, Fermentas Inc., Ontario, CA, USA) and BamHI (10 U/μL, Fermentas Inc.), followed by ligation with the KLF4 target gene, which resulted in a recombinant PLVX-KLF4 plasmid carrying KLF4. Then, the PLVX-KLF4 plasmid was transferred into murine macrophages to construct an upregulated KLF4 cell line (mKLF4).
A KLF4 knockdown vector (KLF4-shRNA) was constructed by RNA interference [15,16]. Following transfection of the lentiviral vector into RAW264.7 cells, stable KLF4 gene silencing was realized in RAW264.7 cells (shKLF4). In addition, cells with the empty plasmid pLVX-mCMV-ZsGreen-PGK-Puro were used as the Negative Control (NC).
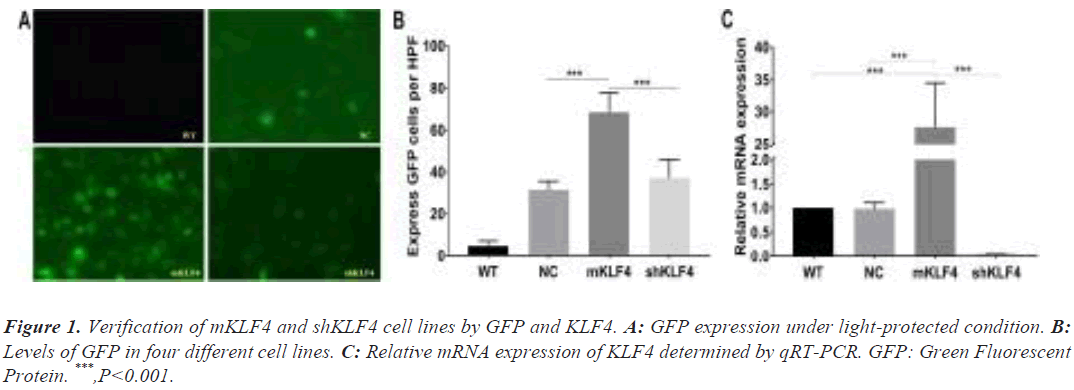
Based on previous findings, cell lines with highly expressed Green Fluorescent Protein (GFP) exhibit high transfection efficiency and proliferative capacity, which suggests successful transfection of pLVX-KLF4 [17]. To verify the successful generation of the mKLF4 and shKLF4 cell lines, GFP expression was observed under light-protected conditions with blue fluorescence.
RNA isolation and real-time PCR analysis
Total RNA was isolated from cultured RAW264.7 macrophages using TRIzol Reagent (CWBIO Co., Ltd.). cDNA was synthesized using a HiFiScript cDNA Synthesis Kit (CWBIO Co., Ltd.). Real-time PCR was conducted with an ABI 7500 RT-PCR system using the SYBR green method. The PCR protocol and sequence information for the PCR primers are described in the Supplement Material.
To evaluate the synergistic effect of KLF4 and GCs on macrophage polarization, we first measured the mRNA levels of signature genes of these two subsets. The iNOS, TNF-α and IL-1β genes have been implicated as mediators of the M1 subset, while Arg-1, TGF-β and IL-10 have been accepted as markers of the M2 subset [18,19].
Protein isolation and western blotting analysis
Macrophages were lysed in Radioimmunoprecipitation Assay (RIPA) buffer with phenylmethylsulfonyl fluoride (PMSF) (Solarbio Co., Ltd.), and the concentration of protein was measured via the Bicinchoninic Acid (BCA) method. Proteins (20 μg/lane) were separated by 10% sodium dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) and transferred to Polyvinylidene Fluoride (PVDF) membranes. Then, PVDF membranes were blocked with 5% nonfat milk at 37°C for 2 h and incubated with primary antibody for 2 h at 37°C or overnight at 4°C. The blots were then washed with Tris- Buffered Saline–Tween 20 (TBST) 3 times and incubated with secondary antibodies for 3 h at 37°C. Bands were detected using ECL Plus enhanced chemiluminescence reagent (Solarbio Co., Ltd.) and quantitated with ImageJ software (version 2.1.4.7 (National Institutes of Health)).
To determine whether there is a synergistic effect of KLF4 and GCs on polarization, CD80 and MHC-II proteins, and CD163 and CD206 proteins were detected as M1 and M2 proteins, respectively, according to recent studies [20,21]. In addition, GR and Histone Deacetylase 2 (HDAC2), crucial downstream signalling molecules in the GR pathway, were used to evaluate the activity of the GR pathway [22,23].
ELISA
After centrifugation, supernatants were harvested and stored at -80ºC. The marker factors levels of M1(IL-6) and M2(Fizz-1) subsets [24] in cell culture supernatants were measured using ELISA kits (Nanjing Jiancheng Bioengineering Institute, Jiangsu, China).
Statistical analysis
Statistical analysis and graph plotting were performed using GraphPad Prism software (version V7.0). Data are presented as the mean ± standard deviation. For comparisons among multiple groups, one-way Analysis Of Variance (ANOVA) followed by Tukey’s post hoc test was used. For those variables with non-normal distributions, the Mann-Whitney test was employed. Two- tailed P-values <0.05 were considered to be significant.
Results
Verification of the mKLF4 and shKLF4 cell lines from the RAW264.7 cell line
The GFP levels of WT, NC, mKLF4 and shKLF4 cells were 4.8 ± 1.07, 31.6 ± 1.81, 68.4 ± 4.17, and 37.2 ± 3.87, respectively. As shown in Figure 1A and Figure 1B, mKLF4 cells expressed the highest GFP levels (mKLF4: WT P<0.001, mKLF4: NC P<0.001, and mKLF4: shKLF4 P<0.001), and there was a significant difference between shKLF4 and WT cells (shKLF4: WT P<0.001).
The KLF4 mRNA levels of WT, NC, mKLF4 and shKLF4 cells measured by qRT-PCR were 1.00 ± 0.00, 0.98 ± 0.07, 27.98 ± 2.04, and 0.05 ± 0.01, respectively (Figure 1C). mKLF4 cells appeared to express the highest KLF4 levels (P<0.001), and shKLF4 cells had lower KLF4 expression than the three other cell lines (P<0.001). No significant difference was found between WT and NC (P=0.786, Figure 1C).
Evaluation of the synergistic polarized regulation via marker mRNA levels
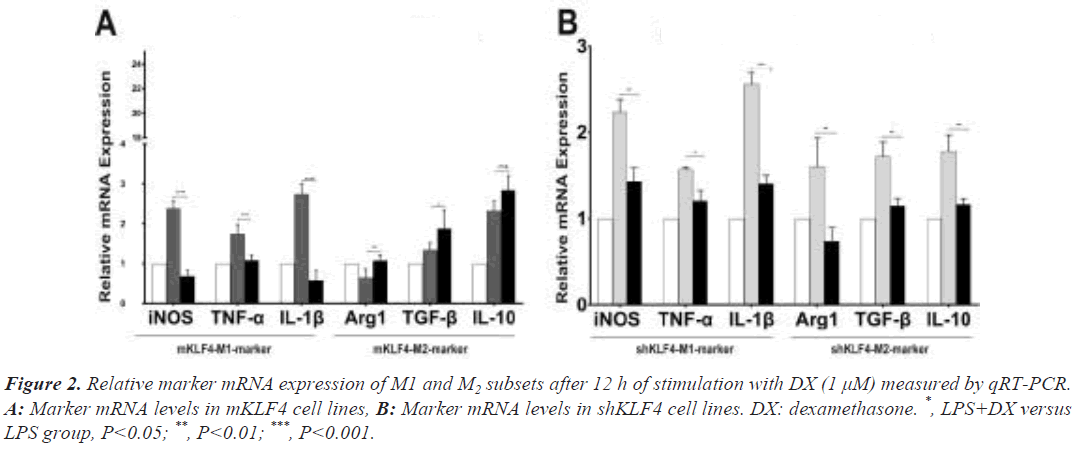
Under regular culture conditions, the mKLF4 group expressed the lowest levels of M1 marker mRNA (M1 mRNA, P<0.001) and the highest levels of M2 marker mRNA (M2 mRNA, P<0.001) compared to levels in the NC group, whereas levels in the shKLF4 cells suggested the opposite trend (Figure S1).
To evaluate the synergistic polarization of KLF4 and GCs, we stimulated mKLF4 and shKLF4 cells with DX and found that M1 mRNA levels were significantly decreased (iNOS: 2.36 ± 0.12 vs. 0.68 ± 0.08, P<0.001; TNF-α: 1.75 ± 0.11 vs. 1.08 ± 0.06, P<0.01; IL-1β: 2.77 ± 0.14 vs. 0.61 ± 0.14, P<0.001) and that M2 mRNA levels were significantly increased (Arg1: 0.65 ± 0.10 vs. 1.08 ± 0.06, P<0.01; TGF-β: 1.35 ± 0.08 vs. 1.879 ± 0.20, P<0.05) in mKLF4 cells (Figure 2A), suggesting that DX and KLF4 played an orchestrated role in driving macrophages towards the M2 phenotype. Further analysis (Figure 2B) of shKLF4 cells demonstrated that M2 mRNA levels were downregulated (Arg1: 1.54 ± 0.15 vs. 0.71 ± 0.07, P<0.01; TGF-β: 1.82 ± 0.12 vs. 1.07 ± 0.08, P<0.01; IL-10: 1.79 ± 0.11 vs. 1.17 ± 0.04, P<0.01) after stimulation with DX, which is consistent with findings observed in mKLF4 cells. Surprisingly, a definite reduction in M1 mRNA levels (iNOS: 2.24 ± 0.08 vs. 1.43 ± 0.09, P<0.01; TNF-α: 1.73 ± 0.16 vs. 1.22 ± 0.05, P<0.05; IL-1β: 2.48 ± 0.10 vs. 1.50 ± 0.10, P<0.001) was detected in shKLF4 cells.
Evaluation of the synergistically polarized regulation via marker protein levels
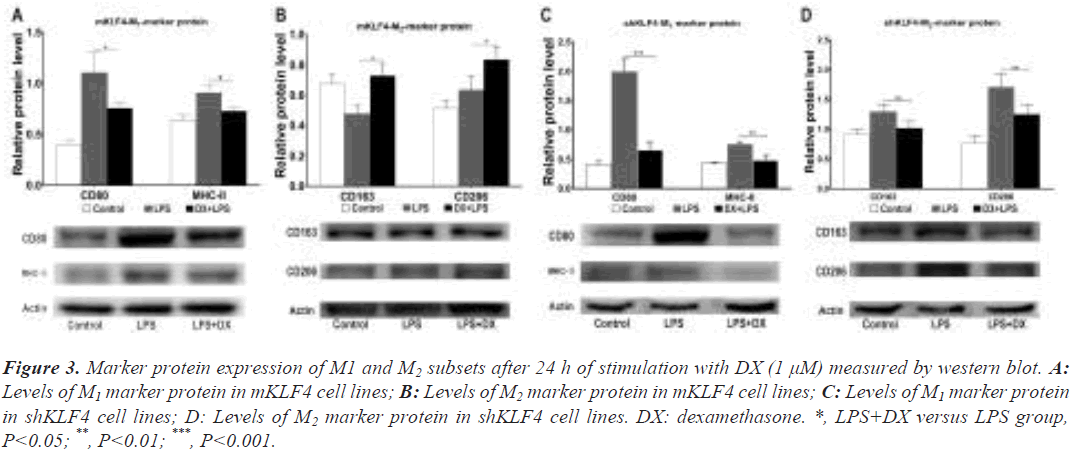
The next section of our trial was concerned with the comparison of M /M marker protein levels. Western blot findings under regular culture conditions revealed a similar trend to that of M1/M2 mRNA levels (P<0.05, Figure S2). To investigate the effect of GCs on polarization regulation at the M1/M2 protein level, DX was added to mKLF4 and shKLF4 cells.
Data from mKLF4 cells revealed that the levels of the M1 proteins CD80 (1.10 ± 0.12 vs. 0.76 ± 0.03, P<0.05) and MHC-II (0.91 ± 0.04 vs. 0.73 ± 0.02, P<0.05) were significantly reduced, while the M2 proteins CD163 (0.48 ± 0.03 vs. 0.73 ± 0.05, P<0.05) and CD206 (0.63 ± 0.05 vs. 0.84 ± 0.05, P<0.05) demonstrated markedly enhanced expression (Figure 3A and Figure 3B). Consistent with the M2 protein results in mKLF4 cells, shKLF4 cells expressed decreased levels of the M2 proteins CD163 (1.30 ± 0.07 vs. 1.02 ± 0.06, P<0.01) and CD206 (1.70 ± 0.13 vs. 1.26 ± 0.09, P<0.01, Figure 3D). Similar to M1 marker mRNA expression in shKLF4 cells, the protein expression of CD80 (1.99 ± 0.12 vs. 0.66 ± 0.08, P<0.001) and MHC- II (0.76 ± 0.02 vs. 0.49 ± 0.05, P<0.01) was significantly downregulated (Figure 3C). These findings also indicated that DX and KLF4 synergistically prime macrophages towards M2 polarization.
Figure 3: Marker protein expression of M1 and M2 subsets after 24 h of stimulation with DX (1 μM) measured by western blot. A: Levels of M1 marker protein in mKLF4 cell lines; B: Levels of M2 marker protein in mKLF4 cell lines; C: Levels of M1 marker protein in shKLF4 cell lines; D: Levels of M2 marker protein in shKLF4 cell lines. DX: dexamethasone. *, LPS+DX versus LPS group, P<0.05; **, P<0.01; ***, P<0.001.
Evaluation of the synergistic polarized regulation via marker factor levels
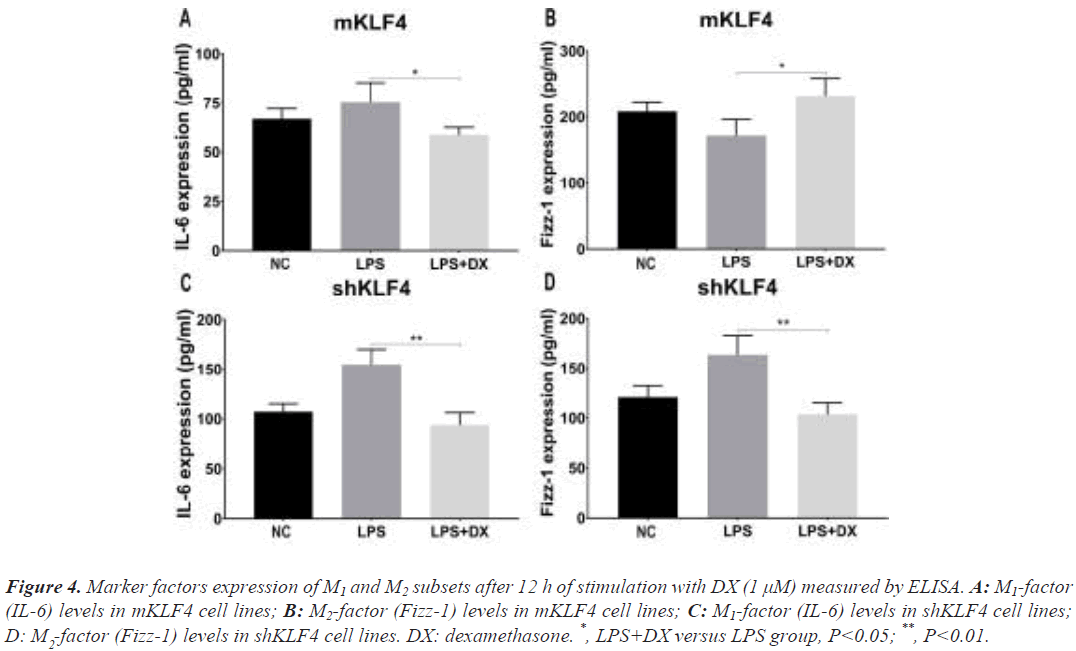
The levels of IL-6 and Fizz-1 under regular culture displayed the same change (Figure S3) as before. After mKLF4 cells were stimulated with DX, M1 factor levels (IL-6) decreased (75.54 ± 5.54 vs. 58.91 ± 2.15, P<0.05, Figure 4A), and M2 factor levels (Fizz-1) significantly increased (171.90 ± 14.01 vs. 231.60 ± 15.25, P<0.05, Figure 4B). Furthermore, the results from shKLF4 cells demonstrated that Fizz-1 levels were reduced (163.50 ± 10.95 vs. 103.60 ± 6.91, P<0.01, Figure 4D). Nevertheless, a definite decrease in IL-6 levels (154.70 ± 8.69 vs. 93.80 ± 7.19, P<0.01, Figure 4C) in shKLF4 cells was found.
Figure 4: Marker factors expression of M1 and M2 subsets after 12 h of stimulation with DX (1 μM) measured by ELISA. A: M1-factor (IL-6) levels in mKLF4 cell lines; B: M2-factor (Fizz-1) levels in mKLF4 cell lines; C: M1-factor (IL-6) levels in shKLF4 cell lines; D: M -factor (Fizz-1) levels in shKLF4 cell lines. DX: dexamethasone. *, LPS+DX versus LPS group, P<0.05; **, P<0.01.
Exploration of the synergistic amelioration of GC resistance by KLF4 and GCs
Western blot results demonstrated that the mKLF4 group contained higher levels of GR (1.29 ± 0.11 vs. 0.89 ± 0.03, P<0.05) and HDAC2 (1.48 ± 0.09 vs. 0.87 ± 0.04, P<0.01) than the levels in the NC group. However, shKLF4 cells exhibited the opposite trend (GR: 0.69 ± 0.07 vs. 0.89 ± 0.03, P<0.05; HDAC2: 0.57 ± 0.06 vs. 0.87 ± 0.04, P<0.01), suggesting that KLF4 could promote the expression of GR pathway proteins, thus mitigating GC resistance (Figure S4).
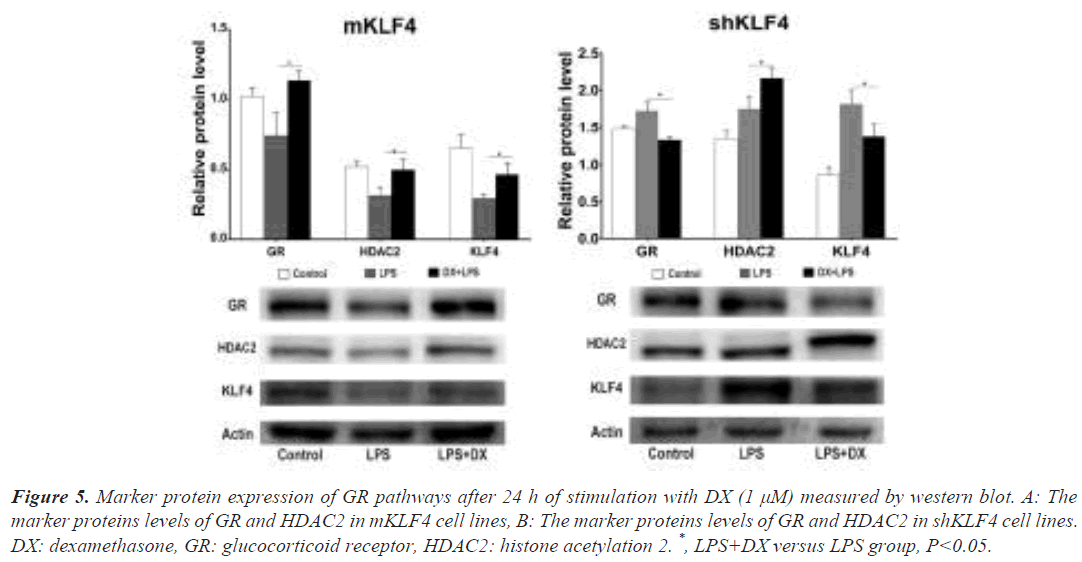
To confirm the synergistic effect of DX and GCs on GR pathways, we measured the expression of GR, HDAC2 and KLF4 in mKLF4 and shKLF4 cells after 24 h of stimulation with DX. As shown in Figure 5A, the protein levels of GR (0.74 ± 0.10 vs. 1.14 ± 0.04, P<0.05), HDAC2 (0.32 ± 0.03 vs. 0.50 ± 0.04, P<0.05) and KLF4 (0.30 ± 0.02 vs. 0.47 ± 0.05, P<0.05) in mKLF4 cells were markedly enhanced compared to those in the LPS (or ILD) group. These findings support that elevated KLF4 and DX cooperate to enhance the expression of GR pathway proteins. Further analysis in shKLF4 cells (Figure 5B) suggested a significant reduction in GR (1.73 ± 0.06 vs. 1.33 ± 0.03, P<0.05) and KLF4 (1.82 ± 0.10 vs. 1.38 ± 0.10, P<0.05), confirming that reduced KLF4 could reduce or even inhibit the expression of GR pathway proteins resulting from DX treatment.
Figure 5: Marker protein expression of GR pathways after 24 h of stimulation with DX (1 μM) measured by western blot. A: The marker proteins levels of GR and HDAC2 in mKLF4 cell lines, B: The marker proteins levels of GR and HDAC2 in shKLF4 cell lines. DX: dexamethasone, GR: glucocorticoid receptor, HDAC2: histone acetylation 2. *, LPS+DX versus LPS group, P<0.05.
Discussion
KLF4, as a critical transcription factor, contributes to inhibiting epithelial-mesenchymal transition in ILD and tissue repair by stimulating M2 polarization of macrophages [10,25]. In this study, KLF4 and GCs cooperated to regulate macrophage polarization towards the M2 subset and to upregulate the expression of GR pathway proteins, thus ameliorating GC resistance.
First, we validated the construction of the mKLF4 and shKLF4 cell lines via GFP and KLF4 mRNA expression levels [26]. The elevation and reduction in GFP in mKLF4 and shKLF4 cells were consistent with our expectation, which illustrates the elevated and decreased transfection efficiency and proliferative capacity in the two cell lines. Additionally, the mRNA expression of KLF4 presented a similar tendency to that of GFP. These data confirmed that macrophages with overexpression and knockdown of KLF4 were successfully constructed. In addition, it is important to mention that the following comparison was performed without the WT group, as the NC and WT groups demonstrated similar expression of KLF4.
With regard to macrophage polarization, mKLF4 cells exhibited higher levels of M2 marker mRNA, protein and factors, whereas shKLF4 cells displayed lower levels of M2 marker mRNA, protein and factors. These results corroborate the ideas of Liao and Saha, who suggested that overexpression and knockdown of KLF4 would promote macrophage polarization towards the M2 and M1 subsets [9,11]. GCs serve as the fundamental therapy in ILD and improve patient outcomes by repressing proinflammatory responses [5]. Previous studies evaluating GCs in ALI observed that GCs ameliorated ALI by promoting M2 polarization [27].
In light of these findings and to explore whether there is a synergistic effect between KLF4 and GCs in macrophage polarization and GC resistance alleviation, we treated mKLF4 and shKLF4 cells in the constructed ILD model with DX. The current study found that M2 marker mRNA, protein and factors were elevated in mKLF4 cells but were reduced in shKLF4 cells after stimulation with DX, which supports the hypothesis that KLF4 and GCs cooperatively polarize macrophages to M2 subsets to promote tissue repair. In addition, the decreased levels of M1 marker mRNA, protein and factors in mKLF4 cells are in accord with that hypothesis. A possible explanation for this might be that GCs inhibit the inflammatory response and regulate polarization via the STAT6 and GR pathways [28,29]. Additionally, the STAT6 pathway serves as one of the key pathways through which KLF4 regulates macrophage polarization [30].
What is surprising is that the levels of M1 marker mRNA, protein and factors in shKLF4 cells were reduced. This inconsistency may be due to the M1 subsets being mainly involved in the inflammatory response, while GCs inhibit inflammation by repressing the activation of inflammatory factors [31,32].
As mentioned previously, DX could promote macrophage polarization by mediating inflammatory factors through GR pathways [29]. Once macrophages are stimulated by external stimuli, the intracellular activated NF-κB pathway gives rise to enhanced histone acetylation, and large amounts of proinflammatory factors (such as TNF-α, IL- 1β, and MMP-9) are released. Under normal conditions, the addition of corticosteroids successively activates GR and upregulates the expression of HDAC2, which can suppress histone acetylation and induce the transformation to the M2 subset. However, abnormal conditions (severe infections, GC resistance, etc.) resulted in suppression of the activity of the GR pathway and enhancement of the activity of the NF-κB pathway even after stimulation with corticosteroids (Figure S5). Ultimately, the burst of inflammatory factors displays an “inflammatory response waterfall” [33,34]. Several lines of evidence suggest that GC resistance is attributed to decreased GR levels, reduced ligand affinity and restricted nuclear translocation [23,35].
Conclusion
Regarding the effect of KLF4 on GR pathways, the up regulated GR and HDAC2 levels in mKLF4 cells under regular culture conditions in our study primarily support that KLF4 could enhance the activity of GR pathways, thus mitigating GC resistance. Furthermore, marker protein expression of GR pathways followed by stimulation with DX was up regulated and down regulated in mKLF4 and shKLF4 cells, respectively. These outcomes confirm that KLF4 and DX cooperate to enhance protein expression of GR pathway members and enhance GR pathway activity, which mitigates GC resistance to some extent. In addition, these findings may provide further support for the hypothesis that KLF4 regulates macrophage polarization via the GR pathway, but this fact remains to be studied.
Our research also has several limitations. First, the primary study was solely conducted in vitro, and we are currently looking at further research using in vivo systems. Second, luciferase assay experiments were not performed to explore whether KLF4 works through the GR-HDAC2 pathway. Last, more studies are needed to explore and validate the mechanism of the cooperative amelioration by GCs and KLF4.
This study validated that KLF4 synergizes with GCs in promoting macrophage polarization towards the M2 subset, which may function via the GR signalling pathway. The second major finding was that KLF4 and GCs act in concert to enhance the expression of GR pathways and ameliorate GC resistance.
Author Contributions
HYS and LS conceived and designed the experiments then DWY and HYS performed the experiments. In addition, HYS and DWY analyzed the data and wrote this paper, finally LQW and LS gave some critical revision of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Natural Science Foundation of Tianjin Province of China (No. 18JCYBJC26600).
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
We would like appreciate our gratitude to Professor Jihong Han in Academy of Life Sciences, Nankai University who donate the RAW264.7 macrophages to us.
References
- Mlika M, Braham E, Mezni F. Interstitial lung disease: Elementary lesions and diagnosis. Semin Diagn Pathol 2018; 35(5):288-296.
[Crossref] [Google Scholar] [PubMed]
- Lescoat A, Lelong M, Jeljeli M, Piquet-Pellorce C, Morzadec C, Ballerie A, Jouneau S, Jego P, Vernhet L, Batteux F. Combined anti-fibrotic and anti-inflammatory properties of JAK-inhibitors on macrophages in vitro and in vivo: Perspectives for scleroderma-associated interstitial lung disease. Biochem Pharmac 2020; 178: 114103.
[Crossref] [Google Scholar] [PubMed]
- Hu X, Xu Q, Wan H, Hu Y, Xing S, Yang H, Gao Y, He Z. PI3K-Akt-mTOR/PFKFB3 pathway mediated lung fibroblast aerobic glycolysis and collagen synthesis in lipopolysaccharide-induced pulmonary fibrosis. Laboratory investigation: A Journal of technical methods and pathology 2020; 100(6): 801-811.
- Zhao J, Yu H, Liu Y, Gibson SA, Yan Z, Xu X, Gaggar A, Li PK, Li C, Wei S. Protective effect of suppressing STAT3 activity in LPS-induced acute lung injury. Am J Physiol Lung cell Mol Physiol 2016; 311(5): L868-L880.
[Crossref] [Google Scholar] [PubMed]
- Van der Goes MC, Jacobs JW, Bijlsma JW. The value of glucocorticoid co-therapy in different rheumatic diseases-positive and adverse effects. Arthritis Res 2014; 16(2): 1-13.
[Crossref] [Google Scholar] [PubMed]
- Oray M, Abu Samra K, Ebrahimiadib N, Meese H, Foster CS. Long-term side effects of glucocorticoids. Expert Opin Drug Saf 2016; 15(4): 457-465.
[Crossref] [Google Scholar] [PubMed]
- Hardy RS, Raza K, Cooper MS. Therapeutic glucocorticoids: mechanisms of actions in rheumatic diseases. Nat Rev Rheumatol 2020; 16(3):133-144.
[Crossref] [Google Scholar] [PubMed]
- Barnes PJ. Corticosteroid resistance in airway disease. Proc Am Thoracic Soc 2004; 1(3):264-268.
[Crossref] [Google Scholar] [PubMed]
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest 2011; 121(7):2736-2749.
[Crossref] [Google Scholar] [PubMed]
- Bialkowska AB, Yang VW, Mallipattu SK. Krüppel-like factors in mammalian stem cells and development. Development (Cambridge, England) 2017; 144(5):737-754.
[Crossref] [Google Scholar] [PubMed]
- Saha B, Bala S, Hosseini N, Kodys K, Szabo G. Krüppel-like factor 4 is a transcriptional regulator of M1/M2 macrophage polarization in alcoholic liver disease. J Leukoc Biol 2015; 97(5):963-973.
[Crossref] [Google Scholar] [PubMed]
- Sevilla LM, Latorre V, Carceller E, Boix J, Vodak D, Mills IG, Perez P. Glucocorticoid receptor and Klf4 co-regulate anti-inflammatory genes in keratinocytes. Mol Cell Endocrinol 2015; 412:281-289.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, Huang Y, Wu C, Du Y, Li P, Wang M, Wang X, Wang Y, Hao Y, Wang T. Network Pharmacology Based Research on the Combination Mechanism between Escin and Low Dose Glucocorticoids in Anti-rheumatoid Arthritis. Front Pharmacol 2019; 10:280.
[Crossref] [Google Scholar] [PubMed]
- Yu X, Zhang R, Lei L, Song Q, Li X. High drug payload nanoparticles formed from dexamethasone-peptide conjugates for the treatment of endotoxin-induced uveitis in rabbit. Int J Nanomedicine 2019; 14:591-603.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Li C, Huang Y, Zhao S, Xu Y, Chen Y, Jiang F, Tao L, Shen X. EOFAZ inhibits endothelial‑to‑mesenchymal transition through downregulation of KLF4. Int J Mol Med 2020; 46(1):300-310.
[Crossref] [Google Scholar] [PubMed]
- Sun L, Li X, Ji W, Wei L. Knockdown of Kruppel like factor 4 (KLF4) promotes RAW264.7 macrophages into M1 polarization. Xi bao yu fen zi mian yi xue za zhi 2020; 36(9):782-787.
[PubMed]
- Ji W, Chen X, Ma Y, Hu D, Lu R, Zhou X, Wei L. Effect of Kruppel like factor 4 gene silencing on apoptosis and phagocytosis of murine RAW264.7 macrophages. Xi bao yu fen zi mian yi xue za zhi 2013; 29(4):341-344.
- Kawano A, Ariyoshi W, Yoshioka Y, Hikiji H, Nishihara T, Okinaga T. Docosahexaenoic acid enhances M2 macrophage polarization via the p38 signaling pathway and autophagy. J Cell Biochem 2019; 120(8): 12604-12617.
[Crossref] [Google Scholar] [PubMed]
- Kang S, Park SJ, Lee AY, Huang J, Chung HY, Im DS. Ginsenoside Rg(3) promotes inflammation resolution through M2 macrophage polarization. J Ginseng Res 2018; 42(1):68-74.
[Crossref] [Google Scholar] [PubMed]
- Dong D, Chen C, Hou J, Yang K, Fang H, Jiang H, Guo F, Wu X, Chen X. KLF4 upregulation is involved in alternative macrophage activation during secondary Echinococcus granulosus infection. Parasite Immunol 2019; 41(10):e12666.
[Crossref] [Google Scholar] [PubMed]
- Lee C, Jeong H, Lee H, Hong M, Park SY, Bae H. Magnolol Attenuates Cisplatin-Induced Muscle Wasting by M2c Macrophage Activation. Front Immunol 2020; 11:77.
[Crossref] [Google Scholar] [PubMed]
- Dschietzig T, Bartsch C, Baumann G, Stangl K. RXFP1-inactive relaxin activates human glucocorticoid receptor: further investigations into the relaxin-GR pathway. Regul Pept 2009; 154(1-3):77-84.
[Crossref] [Google Scholar] [PubMed]
- Jiang Z, Zhu L. Update on molecular mechanisms of corticosteroid resistance in chronic obstructive pulmonary disease. Pulm pharmacol ther 2016; 37:1-8.
[Crossref] [Google Scholar] [PubMed]
- Gensel JC, Zhang B. Macrophage activation and its role in repair and pathology after spinal cord injury. Brain Res 2015; 1619.
[Crossref] [Google Scholar] [PubMed]
- Lin L, Han Q, Xiong Y, Li T, Liu Z, Xu H, Wu Y, Wang N, Liu X. Krüpple-like-factor 4 Attenuates Lung Fibrosis via Inhibiting Epithelial-mesenchymal Transition. Scientific reports 2017; 7(1):15847.
- Gurusinghe S, Bandara N, Hilbert B, Trope G, Wang L, Strappe P. Lentiviral vector expression of Klf4 enhances chondrogenesis and reduces hypertrophy in equine chondrocytes. Gene 2019; 680:9-19.
[Crossref] [Google Scholar] [PubMed]
- Tu GW, Shi Y, Zheng YJ, Ju MJ, He HY, Ma GG, Hao GW, Luo Z. Glucocorticoid attenuates acute lung injury through induction of type 2 macrophage. J Trans Med 2017; 15(1):181.
[Crossref] [Google Scholar] [PubMed]
- Ng Kuet Leong N, Brombacher F, Dalpke AH, Weitnauer M. Crosstalk between glucocorticoids and IL-4 modulates Ym1 expression in alternatively activated myeloid cells. Immunobiology 2017; 222(5):759-767. [Crossref] [Google Scholar] [PubMed]
- Vitellius G, Trabado S, Bouligand J, Delemer B, Lombès M. Pathophysiology of Glucocorticoid Signaling. Ann Endocrinol 2018; 79(3):98-106.
[Crossref] [Google Scholar] [PubMed]
- Porta C, Riboldi E, Ippolito A, Sica A. Molecular and epigenetic basis of macrophage polarized activation. Semin Immunol 2015; 27(4):237-248.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Wang R, Wang H, Yang X, Yang J, Xiong W, Wen Q, Ma L. Glucocorticoids Suppress Antimicrobial Autophagy and Nitric Oxide Production and Facilitate Mycobacterial Survival in Macrophages. Scientific reports 2017; 7(1):982.
- Wang R, Chen W, Ma Z, Li L, Chen X. M1/M2 macrophages and associated mechanisms in congenital bicuspid aortic valve stenosis. Experimental and therapeutic medicine 2014; 7(4):935-940.
[Crossref] [Google Scholar] [PubMed]
- Barnes PJ. Glucocorticosteroids. Handbook of experimental pharmacology 2017; 237:93-115.
[Crossref] [Google Scholar] [PubMed]
- Vandewalle J, Luypaert A, De Bosscher K, Libert C. Therapeutic Mechanisms of Glucocorticoids. Trends Endocrinol Metab 2018; 29(1):42-54.
[Crossref] [Google Scholar] [PubMed]
- Meijer OC, Koorneef LL, Kroon J. Glucocorticoid receptor modulators. Ann Endocrinol 2018; 79(3):107-111.
[Crossref] [Google Scholar] [PubMed]