ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 22
MicroRNA-331-3p promotes cell proliferation and invasion in breast cancer by targeting SRCIN1
Yonghua Hu1#, Wenjun Liu2#, Che Chen1 and Aihua Hou3*
1Department of Internal Medicine, School of Clinical Medicine, GanSu university of Chinese Medicine, Lanzhou, China
2Department of Breast Surgery, Shanxian County Central Hospital of Heze City, Shandong Province, Heze City, Shandong Province, PR China
3Department of Oncology, Yantai City of Shandong Province Hospital of Traditional Chinese Medicine, Yantai City, Shandong Province, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Aihua Hou
Department of Oncology
Yantai city of Shandong Province Hospital of Traditional Chinese Medicine, PR China
Accepted on June 23, 2017
Breast cancer is the most common type of cancer and the most frequent cause of cancer deaths among women worldwide. MicroRNAs (miRNAs) are often dysregulated in various types of human cancer and can function as tumour suppressors or oncogenes in tumourigenesis. MiRNA-331-3p (miR-331-3p) plays an important role in hepatocellular carcinoma, colorectal cancer and glioblastoma; however, its expression level, roles and underlying mechanism in breast cancer remain unknown. Thus, this study investigated the expression level, biological roles and underlying mechanism of miR-331-3p in breast cancer. Results showed that miR-331-3p was significantly up-regulated in breast cancer tissues and cell lines. Down-regulation of miR-331-3p expression inhibited the proliferation and invasion of breast cancer cells. Src kinase signalling inhibitor 1 was validated as a novel direct target of miR-331-3p. Our findings revealed that miR-331-3p might serve as an oncogene, and its over-expression in tumour tissues might contribute to the carcinogenesis and progression of breast cancer. Thus, miR-331-3p is potentially a new therapeutic target for the treatment of breast cancer.
Keywords
Breast cancer, MicroRNA-331-3p, SRCIN1, Proliferation, Invasion.
Introduction
Breast cancer is the most common type of cancer and the most frequent cause of cancer deaths among women worldwide. Approximately 1.7 million diagnoses and about 521,900 deaths, which accounted for 25% of cancer cases and 15% of cancer-related deaths, were reported in 2012 [1]. The incidence of breast cancer is predicted to increase with demographic trend towards an older population, environmental changes and improved diagnostic techniques [2]. Current primary treatments for breast cancer include surgery, hormonal therapy, cytotoxic chemotherapy, immunotherapy and targeted therapy [3]. Despite the great progress in the early diagnosis and systemic therapy of patients with breast cancer, recurrence and distant metastasis remain as major barriers in the successful treatment of breast cancer [4,5]. Therefore, a comprehensive understanding of the molecular pathogenesis of breast cancer is important to develop effective therapeutic measures that are target specific and personalized improve patient survival and prevent metastasis and recurrence.
MicroRNAs (miRNAs) are small, single-stranded, non-coding RNAs that regulate gene expression by interacting preferentially with the 3’ untranslated regions (3’-UTRs) of target mRNAs, which might result in either inhibition of the target protein translation or degradation of the target mRNA [6,7]. MiRNAs and their target mRNAs form complex regulatory networks that are involved in cell proliferation, apoptosis, differentiation stress responses and other biological processes [8,9]. The aberrant expression of miRNAs also contributes to a range of human pathologies, including cancer [10]. Recent studies have found that miRNAs are deregulated in various types of human cancer [11-13]. Depending on the characteristic of their target mRNAs, miRNAs may serve as either oncogenes or tumour suppressors and thus play important roles in tumourigenesis and tumour development [14]. Therefore, miRNAs are potential biomarkers for the diagnosis, treatment and prognosis of human malignancies [15-17].
MiR-331-3p plays an important role in hepatocellular carcinoma [18], colorectal cancer [19] and glioblastoma [20].
However, the expression level, roles and underlying mechanism of miR-331-3p in breast cancer remain unknown. Thus, this study investigated the expression level, biological roles and underlying mechanism of miR-331-3p in breast cancer.
Materials and Methods
Tissue samples
Forty-six paired breast cancer tissues and their adjacent normal tissues were collected from patients who treated with surgery at Shanxian County Central Hospital of Heze City. None patients underwent chemotherapy or radiotherapy prior to surgery. All tissue specimens were immediately frozen in the liquid nitrogen and stored at -80°C refrigerator until further use. This study was approved by Ethical Committee of Shanxian County Central Hospital of Heze City, and written informed consent was provided by each patient.
Cell lines, culture condition and cell transfection
Breast cancer cell lines (MCF-7A, MDA-MB-231, MDA-MB- 453, BT-474, SK-BR-3), a normal mammary epithelial cell line (MCF 10A) and HEK293T cell line were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY), supplemented with 10% Fetal Bovine Serum (FBS; Gibco, Grand Island, NY), 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were grown at 37°C with 5% CO2. All transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
MiR-331-3p inhibitor and corresponding scramble miRNA inhibitor negative control (NC inhibitor) were purchased from Ribobio Technology Co., Ltd. (Guangzhou, China). SRCIN1 overexpression plasmid (pcDNA3.1-SRCIN1) and empty plasmid (pcDNA3.1) were synthesized by GenePharma (Shanghai, China). For transfection, cells were seeded into 6- well plates at a density of 60%-70% confluence. Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol.
Total RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). To detect miR-331-3p expression, reverse transcription was performed using TaqManm MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), followed by qPCR with TaqMan® microRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). U6 was used as an internal control for miR-331-3p expression. For quantitative analysis of SRCIN1 mRNA, cDNA was synthesized using PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). The qPCR was performed using SYBR® Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) on Applied Biosystems 7500 Real-time PCR System (Applied Biosystems, CA, USA), with β-actin as an internal control. All reactions were performed in triplicate and the relative expression of miR-331-3p and SRCIN1mRNA was calculated using the 2-ΔΔCt method.
Cell counting kit 8 (CCK8) assay
Cell proliferation was assessed using the CCK8 (Dojindo, Kumamoto, Japan) assay. After transfection 24 h, cells were seeded into 96-well plates at a density of 3000 cells/well. At various time points following incubation at 37°C, CCK8 assay was performed by adding 10 μl CCK8 reagent into each well. After incubation at 37°C in a 5% CO2 humidified incubator for additional 2 h, cell proliferation was determined by detecting the absorbance at a wavelength of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate and repeated at least three times.
Cell invasion assay
Transwell chambers (8 μm; Corning Incorporated, Corning, NY, USA) coated with Matrigel (BD Biosciences, San Jose, CA, USA) were used to investigate the capacities of cell invasion. Transfected cells were collected at 48 h post-transfection and suspended in FBS-free DMEM. 5 × 104 cells were seeded into the upper chamber, and DMEM containing 20% FBS was placed into the lower chamber as a chemoattractant. After incubation at 37°C in a 5% CO2 humidified incubator for 24 h, cells remaining on the membranes of the transwell chamber were removed carefully with cotton swabs. Invasive cells were fixed in 90% ethanol (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), stained with 0.1% crystal violet (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany),washed with PBS (HyClone, Logan, UT, USA) and dried in air. Cell invasion abilities were evaluated by counting five fields per membrane under an IX51 inverted microscope (Olympus Corporation, Tokyo, Japan; magnification, X200).
Identification of the targets of miR-331-3p
To identify the putative target genes of miR-331-3p, public available bioinformatics tools, TargetScan (http://targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do/), were used to predict the candidate genes.
Luciferase reporter assay
For the luciferase reporter assay, pGL3-SRCIN1-3'UTR wild type (Wt) and pGL3-SRCIN1-3'UTR mutant (Mut) were designed and synthesized by GenePharma. Cells were plated in 24 well plates with 70-80% confluence. After incubation overnight, cells were transfected with miR-331-3p inhibitor or NC inhibitor, followed by co-transfection with pGL3- SRCIN1-3'UTR Wt or pGL3-SRCIN1-3'UTR Mut using Lipofectamine 2000, according to the manufacturer’s protocol. 48 h after transfection, the luciferase activity was determined using the Dual Luciferase Assay System (Promega, Madison, WI, USA), according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Western blotting analysis
Total protein was extracted form tissues or cells with cold radioimmunoprecipitation assay lysis buffer containing protease inhibitors (Beyotime Biotechnology Inc., Shanghai, China). BCA assay kit (Beyotime Biotechnology Inc., Shanghai, China) was used to quantify protein concentration. Equal amounts of protein were separated by 10% Sodium Dodecyl Sulfate (SDS)-polyacrylamide gel electrophoresis gel, transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), and blocked in Tris-buffered saline with Tween-20 (TBST) containing 5% non-fat milk. The membranes were then incubated with primary antibodies against human SRCIN1 (1:1,000 dilution; catalog no.3757; Cell Signaling Technology, USA) or GAPDH (1:1,000 dilution; catalog no.sc-47724; Santa Cruz Biotechnology, CA, USA), at 4°C overnight. After being washed in TBST for three times, the membranes were incubated with corresponding horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; Santa Cruz Biotechnology, CA, USA) at room temperature for 1 h. The proteins bands were visualized by using an enhanced chemiluminescence solution (Pierce; Thermo Fisher Scientific, Inc.) and analysed with AlphaEase FC 4.0.1 software ProteinSimple, San Jose, CA, USA). GAPDH served as an internal control.
Statistical analysis
Data are expressed as mean ± Standard Deviation (SD) and compared with Student's t-test or one-way ANOVA by using the SPSS 19.0 software package (SPSS Inc., Chicago, IL, USA). The association between miR-331-3p and SRCIN1 mRNA expression was analysed using Spearman's correlation analysis. P<0.05 was considered as statistically significant.
Results
MiR-331-3p is highly expressed in breast cancer tissues and cell lines
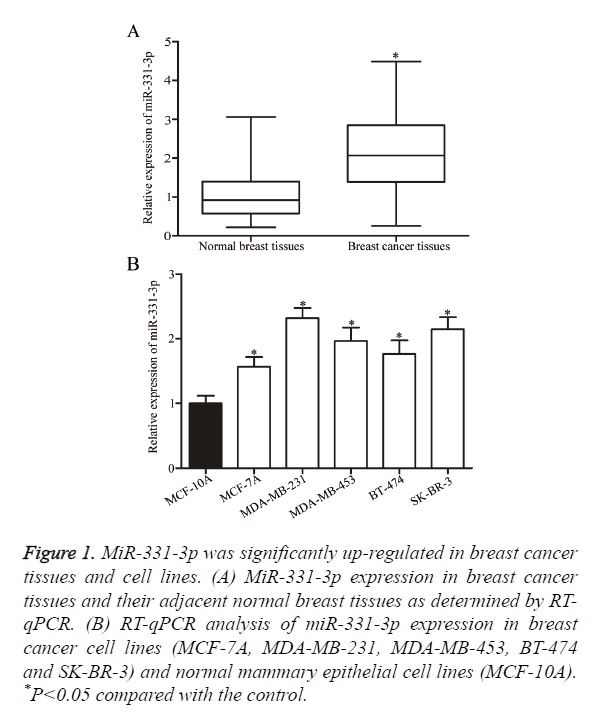
The role of miR-331-3p in breast cancer was investigated by measuring miR-331-3p expression in breast cancer tissues and their adjacent normal breast tissues through RT-qPCR. As shown in Figure 1A, the expression levels of miR-331-3p were significantly higher in breast cancer tissues than in their adjacent normal breast tissues (P<0.05). MiR-331-3p expression was detected in breast cancer cell lines (MCF-7A, MDA-MB-231, MDA-MB-453, BT-474 and SK-BR-3) and normal mammary epithelial cell lines (MCF-10A). Similar to the expression pattern in breast cancer tissues, miR-331-3p was up-regulated in all examined breast cancer cell lines compared with that in MCF-10A (Figure 1B, P<0.05). These results suggest that the deregulated miR-331-3p may play important roles in the initiation and progression of breast cancer.
Figure 1. MiR-331-3p was significantly up-regulated in breast cancer tissues and cell lines. (A) MiR-331-3p expression in breast cancer tissues and their adjacent normal breast tissues as determined by RT-qPCR. (B) RT-qPCR analysis of miR-331-3p expression in breast cancer cell lines (MCF-7A, MDA-MB-231, MDA-MB-453, BT-474 and SK-BR-3) and normal mammary epithelial cell lines (MCF-10A). *P<0.05 compared with the control.
Down-regulation of miR-331-3p expression inhibits the proliferation and invasion of breast cancer cells
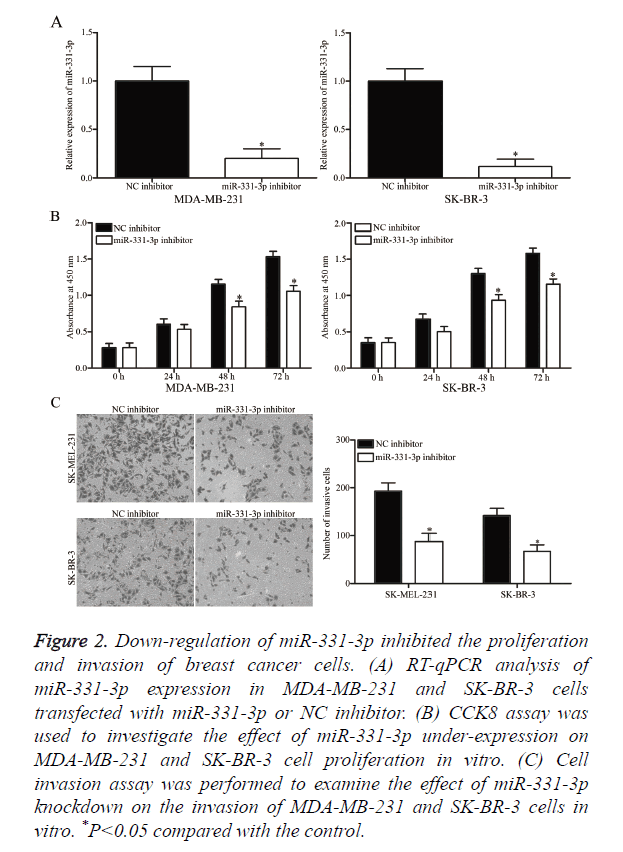
MiR-331-3p or NC inhibitor was transfected into MDA-MB- 231 and SK-BR-3 cells to determine whether miR-331-3p contributes to the formation and progression of breast cancer. At 48 h post-transfection, RT-qPCR analysis confirmed that miR-331-3p expression was down-regulated in MDA-MB-231 and SK-BR-3 cells following transfection with miR-331-3p inhibitor (Figure 2A, P<0.05). CCK8 and cell invasion assays were performed to evaluate the effects of miR-331-3p under expression on the proliferation and invasion of breast cancer cells, respectively. CCK8 assay revealed that down-regulation of miR-331-3p expression inhibited the proliferation of MDA-MB- 231 and SK-BR-3 cells (Figure 2B, P<0.05). Cell invasion assay indicated that miR-331-3p knockdown decreased the invasion of MDA-MB-231 and SK-BR-3 cells (Figure 2C, P<0.05). These results suggest that miR-331-3p serves as an oncogene in breast cancer.
Figure 2. Down-regulation of miR-331-3p inhibited the proliferation and invasion of breast cancer cells. (A) RT-qPCR analysis of miR-331-3p expression in MDA-MB-231 and SK-BR-3 cells transfected with miR-331-3p or NC inhibitor. (B) CCK8 assay was used to investigate the effect of miR-331-3p under-expression on MDA-MB-231 and SK-BR-3 cell proliferation in vitro. (C) Cell invasion assay was performed to examine the effect of miR-331-3p knockdown on the invasion of MDA-MB-231 and SK-BR-3 cells in vitro. *P<0.05 compared with the control.
SRCIN1 is a direct target of miR-331-3p in breast cancer
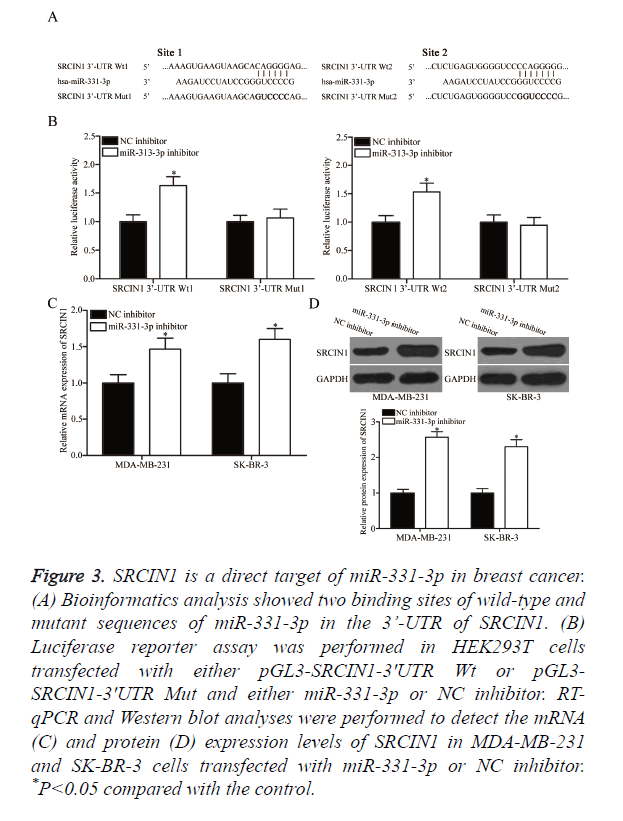
The underlying molecular mechanism of the tumourigenic property of miR-331-3p in breast cancer was explored by predicting the potential target genes through bioinformatics analysis. Among the putative targets, SRCIN1 was selected for further confirmation (Figure 3A) because this gene is lowly expressed in breast cancer and contributes to the formation and progression of breast cancer [21]. Two high-scoring binding sites of miR-331-3p were found on the 3’-UTR of SRCIN1 mRNA. Luciferase reporter assay was conducted in HEK293T cells co-transfected with both miR-331-3p or NC inhibitor and pGL3-SRCIN1-3ʹUTR Wt (1 and 2) or pGL3-SRCIN1-3ʹUTR Mut (1 and 2) to confirm whether SRCIN1 is a direct target of miR-331-3p. Under-expressing miR-331-3p significantly improved the luciferase activity of pGL3-SRCIN1-3ʹUTR Wt (1 and 2; Figure 3B, P<0.05) but exerted no effect on the activity of pGL3-SRCIN1-3ʹUTR Mut (1 and 2). SRCIN1 expression in MDA-MB-231 and SK-BR-3 cells transfected with miR-331-3p or NC inhibitor was measured through RTqPCR and Western blot analyses to determine whether this expression is regulated by miR-331-3p. As shown in Figures 3C and 3D, miR-331-3p inhibitor treatment significantly enhanced the mRNA (P<0.05) and protein (P<0.05) expression levels of SRCIN1 in MDA-MB-231 and SK-BR-3 cells compared with those in the cells transfected with NC inhibitor. These results suggest that SRCIN1 is a direct target of miR-331-3p in breast cancer.
Figure 3. SRCIN1 is a direct target of miR-331-3p in breast cancer. (A) Bioinformatics analysis showed two binding sites of wild-type and mutant sequences of miR-331-3p in the 3’-UTR of SRCIN1. (B) Luciferase reporter assay was performed in HEK293T cells transfected with either pGL3-SRCIN1-3ʹUTR Wt or pGL3- SRCIN1-3ʹUTR Mut and either miR-331-3p or NC inhibitor. RT-qPCR and Western blot analyses were performed to detect the mRNA (C) and protein (D) expression levels of SRCIN1 in MDA-MB-231 and SK-BR-3 cells transfected with miR-331-3p or NC inhibitor. *P<0.05 compared with the control.
SRCIN1 is associated with the effects of miR-331-3p in breast cancer cells
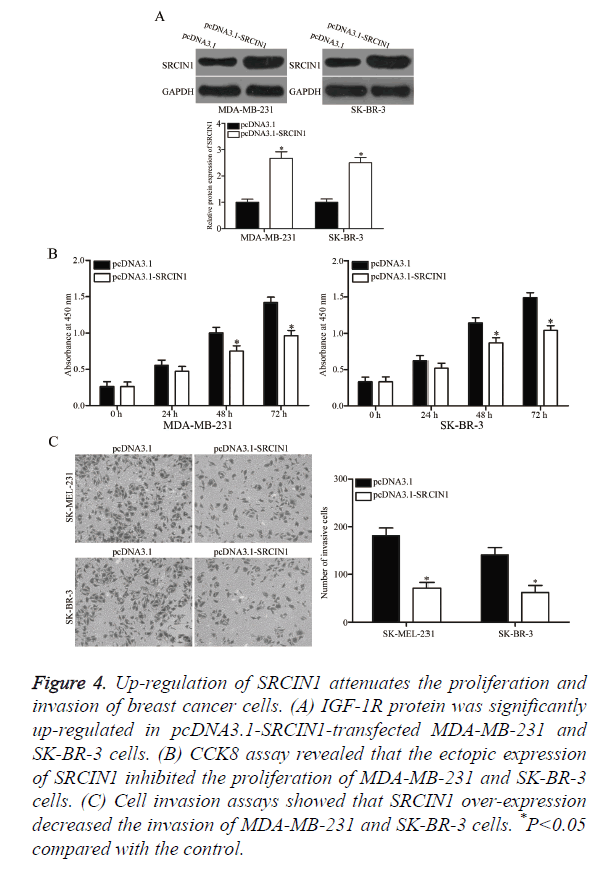
pcDNA3.1-SRCIN1 blank pcDNA3.1 plasmids were transfected into MDA-MB-231 and SK-BR-3 cells to verify whether SRCIN1 is an important mediator of the effects of miR-331-3p in breast cancer cells. As shown in Figure 4A, SRCIN1 was significantly up-regulated in MDA-MB-231 and SK-BR-3 cells after transfection with pcDNA3.1-SRCIN1 plasmid (P<0.05). CCK8 and cell invasion assays revealed that up-regulation of SRCIN1 inhibited the proliferation (Figure 4B, P<0.05) and invasion (Figure 4C, P<0.05) of MDA-MB-231 and SK-BR-3 cells compared with the cells transfected with blank pcDNA3.1 plasmid. These results suggest that the roles of SRCIN1 are similar to those induced by miR-331-3p knockdown in breast cancer cells, thus indicating that SRCIN1 is a functional downstream target of miR-331-3p in vitro.
Figure 4. Up-regulation of SRCIN1 attenuates the proliferation and invasion of breast cancer cells. (A) IGF-1R protein was significantly up-regulated in pcDNA3.1-SRCIN1-transfected MDA-MB-231 and SK-BR-3 cells. (B) CCK8 assay revealed that the ectopic expression of SRCIN1 inhibited the proliferation of MDA-MB-231 and SK-BR-3 cells. (C) Cell invasion assays showed that SRCIN1 over-expression decreased the invasion of MDA-MB-231 and SK-BR-3 cells. *P<0.05 compared with the control.
Discussion
MiR-331-3p is abnormally expressed in many types of malignancies. For example, miR-331-3p is up-regulated in the serum, tissues and cell lines of hepatocellular carcinoma [22,23]. The expression level of miR-331-3p in tissues is highly correlated with the metastasis of hepatocellular carcinoma [22], whereas serum miR-331-3p is associated with TNM stage [23]. High miR-331-3p expression is correlated with the poor long-term survival of patients with hepatocellular carcinoma [22,23]. However, miR-331-3p is lowly expressed in colorectal cancer tissues and cell lines compared with that in healthy colon tissues and cells [19]. MiR-331-3p is also down-regulated in prostate cancer [24], gastric cancer [25] and glioblastoma [20]. These findings suggest that the expression pattern of miR-331-3p is tissue specific; thus, miR-331-3p can be used as a prognostic marker in human cancer.
MiR-331-3p deregulation contributes to the malignant phenotype of several types of human cancer. For instance, Chang et al. reported that miR-331-3p acts as an oncogene in hepatocellular carcinoma by promoting cell growth and metastasis both in vitro and in vivo [22]. However, in gastric cancer, the ectopic expression of miR-331-3p blocks G1/S transition and inhibits colony formation and growth [25]. Zhao et al. [19] reported that the resumed expression of miR-331-3p attenuates cell proliferation, promotes apoptosis and activates caspase-3 in colorectal cancer. Epis et al. [24] revealed that up-regulation of miR-331-3p blocks the androgen receptor signalling pathway in prostate cancer cells, reduces the activity of an androgen-stimulated, prostate-specific antigen promoter and blocks the expression of a prostate-specific antigen. Epis et al. [20] also revealed that restoration of miR-331-3p expression represses the cell proliferation and clonogenic growth in glioblastoma. Fujii et al. [26] indicated that miR-331-3p re-expression suppresses cervical cancer cell proliferation and induces G2/M phase arrest and apoptosis. These conflicting findings revealed that the biological roles of miR-331-3p are tissue specific and can be explained by the ‘imperfect complementarity’ of the interactions between miR-331-3p and its direct target genes. These findings also suggest that miR-331-3p should be investigated as a potential therapeutic target for the treatment of specific cancers.
We then determined the direct target gene of miR-331-3p in breast cancer to explore the mechanisms by which miR-331-3p under-expression inhibits breast cancer cell growth and metastasis. The targets of miR-331-3p include VHL and PHLPP in hepatocellular carcinoma [18,22], DOHH and ERBB-2 in prostate cancer [24,27,28], E2F1 in gastric cancer [25], HER2 in colorectal cancer [19] and NRP2 in cervical cancer [26]. In the present study, an important molecular association between miR-331-3p and SRCIN1 was observed in breast cancer. Firstly, bioinformatics analysis predicted that SRCIN1 is a putative target of miR-331-3p. Secondly, luciferase reporter assay revealed that miR-331-3p can directly target the 3’-UTR of SRCIN1. RT-qPCR and Western blot analyses revealed that miR-331-3p under-expression enhances SRCIN1 expression at the mRNA and protein levels in breast cancer. Moreover, the roles of SRCIN1 over-expression are similar to the effects induced by miR-331-3p knockdown, suggesting that miR-331-3p is a direct functional target of miR-331-3p in breast cancer.
SRCIN1, which is also known as p140 cas-associated protein [29], contains two coiled-coil domains, two proline-rich regions and two regions of highly charged amino acids. According to the characteristic domain structure, SRCIN1 acts as an adaptor protein [30]. SRCIN1 expression is down-regulated in several types of human cancer, such as liver cancer [31], breast cancer [21] and osteosarcoma [32]. Functional experiments revealed that SRCIN1 modulates multiple biological processes, such as cell proliferation, apoptosis, migration, metastasis and invasion [33-35]. In the present study, the expression level of SRCIN1 decreased in the tumour tissues and cell lines of breast cancer. SRCIN1 under expression was also involved in the regulation of cell proliferation, colony formation, apoptosis and metastasis in breast cancer [21]. These findings suggest that the miR-331-3p/ SRCIN1 pathway should be investigated as a potential therapeutic strategy to inhibit the rapid growth and metastasis of breast cancer cells.
In conclusion, miR-331-3p was frequently up-regulated in breast cancer and might serve as an oncogene by directly targeting SRCIN1. MiR-331-3p can be used in miRNA-based therapy for the treatment of breast cancer. However, further studies are still required to evaluate the roles of miR-331-3p in vivo and in a clinical context.
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: 359-386.
- Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev 2017; 26: 444-457.
- Rakha EA. Pitfalls in outcome prediction of breast cancer. J Clin Pathol 2013; 66: 458-464.
- Weigelt B, Peterse JL, van t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer 2005; 5: 591-602.
- Alonso DF, Ripoll GV, Garona J, Iannucci NB, Gomez DE. Metastasis: recent discoveries and novel perioperative treatment strategies with particular interest in the hemostatic compound desmopressin. Curr Pharm Biotechnol 2011; 12: 1974-1980.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281-297.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857-866.
- Gaal Z, Olah E. MicroRNA-s and their role in malignant hematologic diseases. Orv Hetil 2012; 153: 2051-2059.
- Lin S, Pan L, Guo S. Prognostic role of microRNA-181a/b in hematological malignancies: a meta-analysis. PLoS One 2013; 8: 59532.
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012; 482: 347-355.
- Cao Q, Liu F, Ji K. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res 2017; 36: 29.
- Zeng Y, Zhu J, Shen D. MicroRNA-205 targets SMAD4 in non-small cell lung cancer and promotes lung cancer cell growth in vitro and in vivo. Oncotarget 2017; 8: 30817-30829.
- Wang H, Li Q, Niu X, Wang G, Zheng S. miR-143 inhibits bladder cancer cell proliferation and enhances their sensitivity to gemcitabine by repressing IGF-1R signaling. Oncol Lett 2017; 13: 435-440.
- Bhardwaj A, Singh S, Singh AP. MicroRNA-based cancer therapeutics: big hope from small RNAs. Mol Cell Pharmacol 2010; 2: 213-219.
- Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem 2009; 55: 623-631.
- Hou J, Lin L, Zhou W. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011; 19: 232-243.
- Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 2013; 57: 840-847.
- Cao Y, Zhang J, Xiong D. Hsa-miR-331-3p inhibits VHL expression by directly targeting its mRNA 3-UTR in HCC cell lines. Acta Biochim Pol 2015; 62: 77-82.
- Zhao D, Sui Y, Zheng X. MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol Rep 2016; 35: 1075-1082.
- Epis MR, Giles KM, Candy PA, Webster RJ, Leedman PJ. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. J Neurooncol 2014; 116: 67-75.
- Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ. MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel. Gene 2017; 600: 21-28.
- Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology 2014; 60: 1251-1263.
- Chen L, Chu F, Cao Y, Shao J, Wang F. Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumour Biol 2015; 36: 7439-7447.
- Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem 2009; 284: 24696-24704.
- Guo X, Guo L, Ji J. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem Biophys Res Commun 2010; 398: 1-6.
- Fujii T, Shimada K, Asano A, Tatsumi Y, Yamaguchi N. MicroRNA-331-3p suppresses cervical cancer cell proliferation and E6/E7 expression by targeting NRP2. Int J Mol Sci 2016; 17.
- Epis MR, Giles KM, Kalinowski FC, Barker A, Cohen RJ, Leedman PJ. Regulation of expression of deoxyhypusine hydroxylase (DOHH), the enzyme that catalyzes the activation of eIF5A, by miR-331-3p and miR-642-5p in prostate cancer cells. J Biol Chem 2012; 287: 35251-35259.
- Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem 2011; 286: 41442-41454.
- Di Stefano P, Cabodi S, Boeri Erba E. P130Cas-associated protein (p140Cap) as a new tyrosine-phosphorylated protein involved in cell spreading. Mol Biol Cell 2004; 15: 787-800.
- Damiano L, Le Devedec SE, Di Stefano P. p140Cap suppresses the invasive properties of highly metastatic MTLn3-EGFR cells via impaired cortactin phosphorylation. Oncogene 2012; 31: 624-633.
- Chen R, Liao JY, Huang J, Chen WL, Ma XJ, Luo XD. Downregulation of SRC kinase signaling inhibitor 1 (SRCIN1) expression By MicroRNA-32 promotes proliferation and epithelial-mesenchymal transition in human liver cancer cells. Oncol Res 2017.
- Wang P, Wang H, Li X, Liu Y, Zhao C, Zhu D. SRCIN1 suppressed osteosarcoma cell proliferation and invasion. PLoS One 2016; 11: 0155518.
- Gao Y, Xue Q, Wang D, Du M, Zhang Y. miR-873 induces lung adenocarcinoma cell proliferation and migration by targeting SRCIN1. Am J Transl Res 2015; 7: 2519-2526.
- Ye L, Wang H, Liu B. miR-211 promotes non-small cell lung cancer proliferation by targeting SRCIN1. Tumour Biol 2016; 37: 1151-1157.
- Xu X, Wang W, Su N. miR-374a promotes cell proliferation, migration and invasion by targeting SRCIN1 in gastric cancer. FEBS Lett 2015; 589: 407-413.