ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 22
miR-200b suppresses host innate immune response against HBV infection by targeting TLR4 signaling pathway
Yazhen Niu1, Ming Gao2*, Na Feng2 and Jingjing Yang2
1Department of Infectious Diseases, Affiliated Hospital of Hebei University, Baoding, Hebei, PR China
2Department of Radiology, Affiliated Hospital of Hebei University, Baoding, Hebei, PR China
- *Corresponding Author:
- Ming Gao
Department of Radiology
Affiliated Hospital of Hebei University
PR China
Accepted date: October 24, 2017
Background: Toll-Like Receptors (TLRs) play critical roles in innate immunity. HBV can modulate the expression of TLRs and/or inhibit TLR signaling cascades to escape the innate immune response. However, the mechanism of HBV in the regulation of TLRs remains unclear. In this study, we aimed to clarify whether miR-200b suppresses host innate immune response against HBV infection by targeting TLR4 signaling pathway.
Methods: First, the expression of miR-200b in HBV-infected liver tissues and HBV-producing HepG2.2.15 cells was detected using Quantitative Real-time PCR (qRT-PCR). We then examined the association between chronic HBV infection and the expression of miR-200b. ELISA assay was used to evaluate the host innate immune response. Further, the potential target genes related to innate immunity of miR-200b were also predicted. Western blot and luciferase reporter assays were performed to confirm the predicted genes.
Results: An upregulated expression of miR-200b was observed in HBV-infected liver tissues and HepG2.2.15 cells. Besides, a positive association was found between chronic HBV infection and the expression of miR-200b. A luciferase reporter assay revealed that MyD88 and IRAK4 were the downstream targets of miR-200b. Finally, knockdown of miR-200b in HepG2.2.15 cells suppressed TLR4-mediated innate response.
Conclusions: Our study revealed that miR-200b was related to HBV infection through a pathway involving the MyD88 and IRAK4 and provides new insights into the understanding of mechanisms of HBV-induced immunosuppressive strategy.
Keywords
miR-200b, Toll-like receptor 4, HBV infection, Innate immunity
Introduction
Hepatitis B Virus (HBV) is an enveloped, non-cytopathic virus that causes variable degrees of liver disease in humans, including acute and chronic necroinflammatory liver diseases and, subsequently, hepatic cirrhosis and hepatocellular carcinoma [1,2]. Despite the access to a highly effective prophylactic vaccine, 350 million people have been infected with HBV worldwide, especially prevalent in Asia and Africa.
The role of innate immunity in defense against HBV was a matter of debate for a number of years. However, several studies have now provided clear evidence that HBV could be sensed by the immune system early after infection, and viral replication and dissemination is controlled by innate responses [3,4]. Alternatively, in order to establish a chronic infection, HBV may evade this innate response or downregulates antiviral pathways, such as Toll-Like Receptors (TLRs) pathway [5-7]. Thus, it is crucial to the advancement of our understanding to the mechanisms of HBV and host innate immunity interaction.
MicroRNAs (miRNAs) are small non-coding RNAs which are key regulators of many cellular events, including cell proliferation, differentiation, immune response and metabolism [8-10]. Accumulating evidence has indicated that abnormal expression of miRNAs is closely associated with many diseases, including liver diseases [11]. Among those miRNAs, miR-200b is reported to be commonly related to epithelial-mesenchymal transition [12], angiogenic response [13], chemoresistance [14] and female fertility [15]. However, whether miR-200b is involved in regulating HBV-induced host innate immune response remains obscure. Here we demonstrate that miR-200b inhibits TLR4-mediated innate immunity by targeting MyD88 and IRAK4 in HBV infection.
Material and Methods
Patients
Sixty-two consecutive HBsAg/anti-HBe/HBV-DNA-positive and HbeAg-negative patients with chronic HBV infection underwent a diagnostic liver biopsy from May 2012 to June 2014. None of them had previously been infected with HCV, HDV or HIV and treated with antiviral or immunosuppressive therapies. Twenty-five healthy controls were negative for HBV, HCV or HIV infection and tested normal for Aspartate Aminotransferase (AST) and Alanine Transaminase (ALT). An informed written consent in compliance with the Helsinki declaration was obtained from all the patients. The study was approved by the Ethics Committee of Affiliated Hospital of Hebei University.
Cell culture and transfection
HepG2 and HepG2.2.15 were cultured in DMEM/High Glucose medium (HyClone, Thermo scientific Inc., China). HEK293T cells were grown in RPMI-1640 medium (GIBCO/ BRL, Grand Island, NY, USA) supplemented with 10% FBS. All cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2. Transfections were performed with LipofectamineTM 2000 Reagent (Invitrogen, CA, USA) following the manufacturer’s protocol.
RNA isolation, reverse transcription, and quantitative Real-time PCR (qRT-PCR)
Total RNA was extracted from the liver tissues and cells using TRIzol reagent (Invitrogen, Karlsruhe, Germany). cDNAs were synthesized using MicroRNA cDNA kit (Exiqon, Woburn, MA). qRT-PCR was performed using an ABI 7900 cycle detection system. The data were analysed via the comparative Ct (2-ΔΔCT) method. The expression of the miRNAs was normalized to U6 and reported as Arbitrary Units (AU).
Detection of the main clinical indicators
HBV DNA was quantified by COBAS TaqMan assay (Roche Diagnostic Systems Inc., Mannheim, Germany). Serum AST and ALT levels were detected using an automatic biochemical analyzer (Hitachi Ltd., Japan).
ELISA assay
The levels of IL-6 and TNF-α in cell culture supernatants were determined using ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instruction.
Western blot
Antibodies directed against IRAK4, MyD88, NF-κB p65 and phosphorylated NF-κB p65 (1:1000, Abcam) were used to determine protein level by Western blot. β-actin (1:2500, Abcam) was detected as loading control.
Luciferase reporter assay
A fragment of the Wild-Type (WT) or Mutated type (MUT) 3’ untranslated region (UTR) of IRAK4 and MyD88 containing the putative miR-200b binding site were cloned into a BamH I/Sal I site of the pGL3-basic luciferase reporter vector (Promega). For the luciferase assay, 293T cells were cotransfected with pGL3-constructs with or without miR-200b mimics. Forty-eight hours after transfection, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity.
Statistical analysis
Experimental data were presented as the mean ± standard deviation with at least three repeats. The statistical significance of differences between groups was evaluated using Student’s ttest and a P-value less than 0.05 was considered statistically significant.
Results
Frequent upregulation of miR-200b in HBV-infected liver tissues and HBV-producing cells
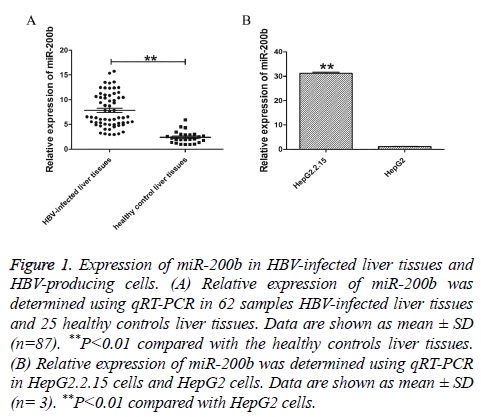
To investigate the potential function of miR-200b in chronic HBV infection, we first sought to examine the expression of miR-200b in 62 samples HBV-infected liver tissues and 25 healthy controls liver tissues using qRT-PCR. The results showed that miR-200b was frequently upregulated in HBV-infected liver issues compared with normal tissues (Figure 1A, P<0.01). In order to further verify the expression level of miR-200b, we analysed the expression of miR-200b in HepG2.2.15 cells and HepG2 cells. As shown in Figure 1B, miR-200b was similarly enriched in HepG2.2.15 cells (P<0.01).
Figure 1: Expression of miR-200b in HBV-infected liver tissues and HBV-producing cells. (A) Relative expression of miR-200b was determined using qRT-PCR in 62 samples HBV-infected liver tissues and 25 healthy controls liver tissues. Data are shown as mean ± SD (n=87). **P<0.01 compared with the healthy controls liver tissues. (B) Relative expression of miR-200b was determined using qRT-PCR in HepG2.2.15 cells and HepG2 cells. Data are shown as mean ± SD (n= 3). **P<0.01 compared with HepG2 cells.
miR-200b expression associated with the severity of liver disease
To explore the potential role of miR-200b in HBV-related liver disease, we further determined miR-200b expression in different stages of HBV infection patients using qRT-PCR. Compared with normal liver tissue, miR-200b expression in the liver tissues taken from patients with chronic HBV infection, HBV-induced cirrhosis and HBV-related hepatocellular carcinoma was 5.78 ± 1.28, 0.57 ± 0.31 and 0.14 ± 0.27 (Table 1).
| Group | Cases | Gender (male/female) | Age range | Average age | miR-200b expression, A.U., mean ± SD | P-value |
|---|---|---|---|---|---|---|
| Healthy controls | 20 | (13/7) | 20-56 | 32.00 ± 7.36 | 1.04 ± 0.67 | |
| Chronic HBV infection | 40 | (30/10) | 12-65 | 38.18 ± 14.87 | 5.78 ± 1.28 | P<0.01 |
| HBV-induced cirrhosis | 35 | (26/9) | 25-68 | 43.05 ± 12.98 | 0.57 ± 0.31 | P<0.05 |
| HBV-related hepatocellular carcinoma | 14 | (10/4) | 28-70 | 48.38 ± 15.34 | 0.14 ± 0.27 | P<0.01 |
| SD: Standard Deviation, Paired t-tests were used for comparing chronic HBV infection, HBV-induced cirrhosis or HBV-related hepatocellular carcinoma to healthy controls, P<0.05 was considered as significant difference. | ||||||
Table 1: miR-200b expression associated with the severity of liver disease.
Correlation of miR-200b with clinical indicators of patients with CHB
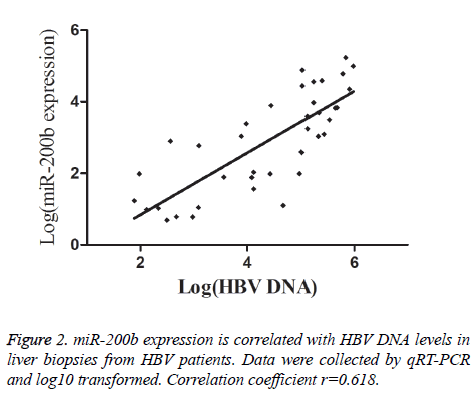
To further analyse whether there is a correlation between miR-200b levels and chronic HBV infection, we checked HBV DNA load in liver biopsies from HBV patients. As shown in Figure 2, there was a direct correlation between miR-200b expression and liver HBV DNA load. miR-200b expression was also analysed according to the patients’ AST and ALT serum values. Significantly higher miR-200b expression was observed in patients with abnormal AST serum levels (mean ± SD: 2.99 ± 2.20 vs. 0.22 ± 0.16, P<0.001), in those with abnormal ALT serum levels (mean ± SD: 4.55 ± 3.15 vs. 0.23 ± 0.18, P<0.001) (Table 2).
| Parameters | Number of patients | miR-200b expression, A.U., mean ± SD | P-value |
|---|---|---|---|
| AST | |||
| ≤ 1 × n.v. | 15 | 0.24 ± 0.12 | |
| >1 × n.v. | 9 | 1.54 ± 0.95 | 0.003 |
| ALT | |||
| ≤ 1 × n.v. | 18 | 0.23 ±0.12 | |
| >1 × n.v. | 10 | 1.28 ±1.01 | 0.009 |
| AST: Aspartate-Aminotransferase; ALT: Alanine-Aminotrasferase; n.v.: Normal Value; SD: Standard Deviation; Comparison of the data sets was performed by Student’s t-test, P<0.05 was considered as significant difference. | |||
Table 2: miR-200b expression according to liver disease.
miR-200b negatively regulated IRAK4 and MyD88
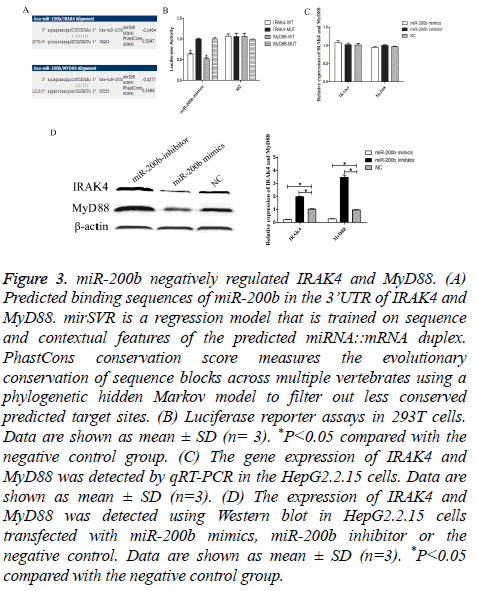
Through bioinformatics software (microRNA.org), we sought to identify its downstream targets and molecular pathways of miR-200b specifically associated with TLR4 pathway. Among the 7,176 mRNA transcripts, IRAK4 and MyD88 were predicted to harbor evolutionarily conserved binding site(s) of miR-200b (Figure 3A). To experimentally validate software prediction results, 3’UTRs of IRAK4 and MyD88 were cloned into a luciferase reporter construct and co-transfected with miR-200b mimics in 293T. We found that co-transfection of miR-200b and pGL3- IRAK4 or MyD88 WT vector significantly decreased the luciferase activity in 293T cells, as compared with the control (P<0.05). However, miR-200b had no effect on the luciferase activity when transfected with pGL3-IRAK4 or MyD88 MUT vector (Figure 3B). We observed a dramatic decrease of IRAK4 and MyD88 protein level in HepG2.2.15 cells transfected with miR-200b mimics compared with the negative control (Figure 3D, P<0.05). Meanwhile, knockdown of miR-200b in HepG2.2.15 cells could obviously increase IRAK4 and MyD88 protein level.
Figure 3: miR-200b negatively regulated IRAK4 and MyD88. (A) Predicted binding sequences of miR-200b in the 3’UTR of IRAK4 and MyD88. mirSVR is a regression model that is trained on sequence and contextual features of the predicted miRNA::mRNA duplex. PhastCons conservation score measures the evolutionary conservation of sequence blocks across multiple vertebrates using a phylogenetic hidden Markov model to filter out less conserved predicted target sites. (B) Luciferase reporter assays in 293T cells. Data are shown as mean ± SD (n= 3). *P<0.05 compared with the negative control group. (C) The gene expression of IRAK4 and MyD88 was detected by qRT-PCR in the HepG2.2.15 cells. Data are shown as mean ± SD (n=3). (D) The expression of IRAK4 and MyD88 was detected using Western blot in HepG2.2.15 cells transfected with miR-200b mimics, miR-200b inhibitor or the negative control. Data are shown as mean ± SD (n=3). *P<0.05 compared with the negative control group.
However, miR-200b can’t effect IRAK4 and MyD88 expression in mRNA level (Figure 3C). These results suggested that IRAK4 and MyD88 were the direct target genes of miR-200b.
miR-200b suppressed MyD88/IRAK4 mediated innate response
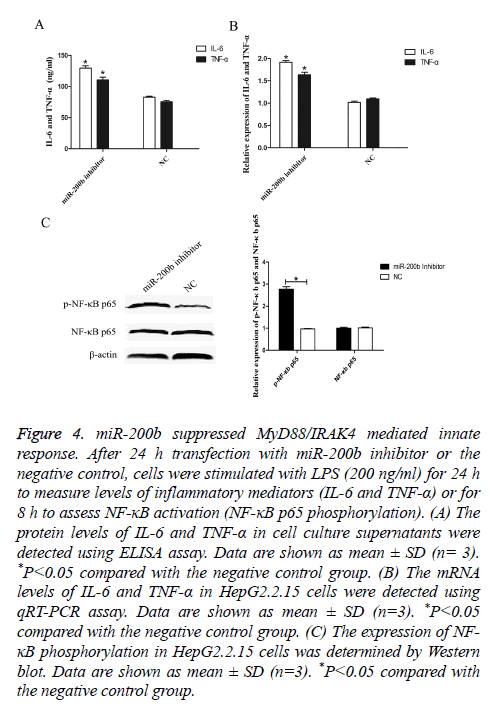
To elucidate the effect of miR-200b on TLR4 pathway mediated innate response, ELISA assay was performed to analyse the expression levels of IL-6 and TNF-α in vitro. After 24 h transfection, cells were stimulated with LPS (200 ng/ml) for 24 h and cell culture supernatants were collected. As shown in Figure 4A, the levels of IL-6 and TNF-α were obviously increased in HepG2.2.15 cells transfected with miR-200b inhibitor compared with the negative control (P<0.05).
Figure 4: miR-200b suppressed MyD88/IRAK4 mediated innate response. After 24 h transfection with miR-200b inhibitor or the negative control, cells were stimulated with LPS (200 ng/ml) for 24 h to measure levels of inflammatory mediators (IL-6 and TNF-α) or for 8 h to assess NF-κB activation (NF-κB p65 phosphorylation). (A) The protein levels of IL-6 and TNF-α in cell culture supernatants were detected using ELISA assay. Data are shown as mean ± SD (n= 3). *P<0.05 compared with the negative control group. (B) The mRNA levels of IL-6 and TNF-α in HepG2.2.15 cells were detected using qRT-PCR assay. Data are shown as mean ± SD (n=3). *P<0.05 compared with the negative control group. (C) The expression of NF- κB phosphorylation in HepG2.2.15 cells was determined by Western blot. Data are shown as mean ± SD (n=3). *P<0.05 compared with the negative control group.
Moreover, similar findings were also observed in the mRNA levels of IL-6 and TNF-α as demonstrated by qRT-PCR (Figure 4B, P<0.05). In addition, knockdown of miR-200b augmented the expression levels of phosphorylated NF-κB p65 compared with the negative control (Figure 4C, P<0.05). These results indicated that miR-200b may promote HBV infection through suppressing MyD88/IRAK4 mediated innate response.
Discussion
Innate immunity represents the first line of defense against HBV. HBV could interact with the host’s innate and adaptive immune responses to establish chronic infection. The study demonstrated that TLRs are essential for the recognition of invading pathogens and serve as an important link between innate and adaptive immunity [16]. TLRs recognize Pathogen- Associated Molecular Patterns (PAMPs) to initiate protective immune responses [17,18]. MyD88 and IRAK4 are critical signaling mediators of the TLR/IL1-R superfamily [19,20]. In humans, inherited MyD88 and IRAK4 deficiencies cause recurrent, often life-threatening infections by pyogenic bacteria [21,22].
By comparing the expression of 1,008 miRNAs between HepG2.2.15 cells and parallel hepatoma cells HepG2, Jiang et al. observed several differential expression of miRNAs related to TLRs pathways, indicating miRNAs during HBV infection might play a critical role in innate immunity against HBV infection [23]. Previous studies have found cellular miRNAs can positively or negatively influence virus replication and regulate HBV-related liver disease, either by targeting cellular transcription factors required for HBV gene expression or by a direct binding to HBV transcripts [24].
In the present study, we demonstrated that frequent up-regulation of miR-200b in chronic HBV-infected liver tissues and HBV-producing cells attenuated TLR4-mediated antiviral responses of innate immunity by targeting its downstream signaling mediators MyD88 and IRAK4. In this way, HBV avoids recognition by the innate immune system and forms a persistent infection. This may explain previous observations on its apparent inability to be detected by PRRs.
Using qRT-PCR, we found a differential expression of miR-200b in different stages of HBV infection patients. Compared with healthy control, there was a significant change of the expression of miR-200b during the development from chronic HBV infection to HBV-induced cirrhosis. This change may result from the change of immune system function caused by HBV infection during the early stage of HBV-related hepatocarcinogenesis. Future studies will be needed to explain the mechanism of how the interactions between replicating HBV and the immune system resulted in the change of miRNAs expression profile.
To conclude, our study revealed upregulation of miR-200b in chronic HBV infection suppresses TLR4-mediated innate immunity by targeting MyD88 and IRAK4 and subsequently promotes HBV infection. We believe that these findings may contribute to the understanding of the pathogenesis of HBV infection and the HBV-related hepatocarcinogenesis.
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgements
We thank all the staff in the department of Infectious Diseases of Affiliated Hospital of Hebei University for their help on the manuscript.
References
- Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet 2014; 384: 2053-63.
- Dienstag JL. Hepatitis B virus infection. N Engl J Med 2008; 359: 1486-1500.
- Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol 2013; 47: 2-6.
- Ait-Goughoulte M, Lucifora J, Zoulim F, Durantel D. Innate antiviral immune responses to hepatitis B virus. Viruses 2010; 2: 1394-1410.
- Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol 2005; 79: 7269-7272.
- Chen Z, Cheng Y, Xu Y, Liao J, Zhang X, Hu Y, Zhang Q, Wang J, Zhang Z, Shen F, Yuan Z. Expression profiles and function of Toll-like receptors 2 and 4 in peripheral blood mononuclear cells of chronic hepatitis B patients. Clin Immunol 2008; 128: 400-408.
- Wu J, Meng Z, Jiang M, Pei R, Trippler M, Broering R, Bucchi A, Sowa JP, Dittmer U, Yang D, Roggendorf M, Gerken G, Lu M, Schlaak JF. Hepatitis B virus suppresses toll-like receptor-mediated innate immune responses in murine parenchymal and nonparenchymal liver cells. Hepatology 2009; 49: 1132-1140.
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006; 11: 441-450.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857-866.
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci 2007; 32: 189-197.
- Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol 2009; 15: 1665-1672.
- Chen Y, Xiao Y, Ge W, Zhou K, Wen J, Yan W, Wang Y, Wang B, Qu C, Wu J, Xu L, Cai W. miR-200b inhibits TGF-β1-induced epithelial-mesenchymal transition and promotes growth of intestinal epithelial cells. Cell Death Dis 2013; 4: 541.
- Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem 2011; 286: 2047-2056.
- Wu Y, Xiao Y, Ding X, Zhuo Y, Ren P, Zhou C, Zhou J. A miR-200b/200c/429-binding site polymorphism in the 3 untranslated region of the AP-2α gene is associated with cisplatin resistance. PLoS One 2011; 6: 29043.
- Hasuwa H, Ueda J, Ikawa M, Okabe M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 2013; 341: 71-73.
- Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Microbes Infect 2004; 6: 1382-1387.
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124: 783-801.
- Beutler B. Inferences, questions and possibilities in toll-like receptor signaling. Nature 2004; 430: 257-263.
- Dunne A, Oneill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE 2003; 2003: 3.
- Bowie A, Oneill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal generators for pro-inflammatory interleukins and microbial products. J Leukoc Biol 2000; 67: 508-514.
- Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, Elbim C, Hitchcock R, Lammas D, Davies G, Al-Ghonaium A, Al-Rayes H, Al-Jumaah S, Al-Hajjar S, Al-Mohsen IZ, Frayha HH, Rucker R, Hawn TR, Aderem A, Tufenkeji H, Haraguchi S, Day NK, Good RA, Gougerot-Pocidalo MA, Ozinsky A, Casanova JL. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 2003; 299: 2076-2079.
- von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, Vasconcelos J, Sirvent N, Guedes M, Vitor AB, Herrero-Mata MJ, Arostegui JI, Rodrigo C, Alsina L, Ruiz-Ortiz E, Juan M, Fortuny C, Yague J, Anton J, Pascal M, Chang HH, Janniere L, Rose Y, Garty BZ, Chapel H, Issekutz A, Maródi L, Rodriguez-Gallego C, Banchereau J, Abel L, Li X, Chaussabel D, Puel A, Casanova JL. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 2008; 321: 691-696.
- Jiang X, Kanda T, Wu S, Nakamura M, Miyamura T, Nakamoto S, Banerjee A, Yokosuka O. Regulation of microRNA by hepatitis B virus infection and their possible association with control of innate immunity. World J Gastroenterol 2014; 20: 7197-7206.
- Kitab B, Alj HS, Ezzikouri S, Benjelloun S. MicroRNAs as important players in host-hepatitis B virus interactions. J Clin Transl Hepatol 2015; 3: 149-161.