ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2008) Volume 19, Issue 1
Molecular characterization of verotoxin gene in enteropathogenic Escherichia coli isolated from Miri Hospital, Sarawak, Malaysia.
Department of Microbiology, Faculty of Medicine, Universiti Teknologi MARA, 40450 Shah Alam, Selangor, Malaysia.
- Corresponding Author:
- Dr. Nazmul
Faculty of Medicine
Universiti Teknologi MARA
Shah Alam, 40450 Selangor
Malaysia
E-mail: poorpiku@yahoo.com
Phone: +60 129771486
Accepted Date: December 7 2007
Thirty enteropathogenic Escherichia coli (EPEC) isolated (Miri hospital, Sarawak, Malaysia, 2003) from children with diarrhea were examined for the detection of verotoxin (VT). PCR method was carried out using two sets of primers (VT1 and VT2) which yielded a fragment of 348 bp of VT1 gene and 584 bp of VT2 gene, respectively. Ten E. coli isolates (33%) carried VT1 gene but none was found to carry VT2 gene. All the VT1-positive isolates harbored plasmid. Southern hybridiza-tion study revealed that VT1 probe was in homology with the chromosome of all 10 VT1-positive isolates indicating that the VT1 gene has to be located on chromosome. Nucleotide sequencing was made from the VT1 gene fragment isolated from an EPEC isolates (E. coli 07). It showed that 96% homology with the known VT1 gene indicates the similar gene harbored by E. coli O157 and other ancestors.
Keywords
Molecular characterization, verotoxin gene, EPEC.
Introduction
The term enteropathogenic Escherichia coli (EPEC) was proposed by Nester in 1955 to differentiate between the incriminated diarrheal Escherichia coli (E. coli) and the normal flora [1]. EPEC is a major cause of diarrhea in infants throughout the world. Clinically, EPEC infections are characterized by fever, malaise, vomiting and diarrhea with prominent amount of mucous but no blood [2]. EPEC strains cause a distinctive ultra structural histopa-thologic lesion in human, a property not shared by non-pathogenic E. coli strains and other classes of pathogenic E. coli. A virulence property that has been associated with EPEC is the production of verotoxin (VT). It was reported that some EPEC strains elaborate moderate quantities of cytotoxin very similar or identical to the toxin VT1 and VT2, which might play a role in the pathogenesis of EPEC disease [3,]. Many of the cytotoxigenic strains re-ferred to as EPEC serogroups and serotyped by Marques et al. [3] and Cleary et al. [4] should be more correctly categorized as enterohemorrhagic E. coli (EHEC). EPEC upon producing VT1 or VT2 was named as EHEC. There are three types of E. coli VTs - VT1, VT2 (human strains) and VT2 variant (porcine strain). Verotoxins, also known as Shiga-like toxin was first identified in Shigella dysenteriae, where it is chromosomally encoded, but the genes for its production are transmitted between E. coli strains by toxin encoding bacteriophages [5]. Both the antigenically distinct toxins VT1 and VT2 are encoded on two different lysogenic lambdoid bacteriophages (933J and 933W) inserted into the chromosome in E. coli 933 [6]. This virulence factor (VT or SLT) is found in many different serotypes. A few well characterized bioserotypes namely O157:H7 and O26:H11 are the most important hosts for the verotoxin phage(s) [7].
The PCR is an easy, rapid, inexpensive protocol that per-mits accurate and detection of important toxin genes in clinical material and possibly in contaminated foods [8]. DNA hybridization technique is very fast and specific. Synthetic oligonucleotide probes from A and B subunits of VT1 and VT2 have been constructed and these probes have been used at different degrees of stringency to iden-tify VT1, VT2 and pig edema variants of VT2 producing E. coli [5].
Materials and Methods
Bacterial strains
Thirty confirmed EPEC isolates (E. coli 01-E. coli 30) were obtained from infants with diarrhea (Hospital of Miri, Sarawak, Malaysia, 2003). E. coli O157:H7 EDL933 (carries VT-1 and VT-2 genes) was used as posi-tive control and E. coli JM101 was used as a negative control. The isolates were cultured onto brain heart infu-sion agar and brain heart infusion broth.
PCR
The VT1 forward, VT1 reverse, VT2 forward and VT2 reverse primers were used for the PCR method. The se-quences for these primers were obtained from Cebula et al. [9]. Expected amplicon size for VT1 is 348 bp, and for VT2 is 584 bp. The primers were synthesized commer-cially from Biosyntech Company, Malaysia. Taq DNA polymerase, dNTP, MgCl2, 10X buffer for PCR were pur-chased from Promega, USA. The parameter and the steps for PCR protocol were followed exactly as described by Nazmul et al. [5].
Plasmid DNA extraction
Plasmids were extracted by the alkaline lysis modified method which was already established by Salmah [10]. Plasmid DNA was resolved by electrophoresis in sub-merged horizontal agarose (Sigma Chemical Co., USA) slab gel (0.7%) in Tris-Acetate buffer (TAE, pH 8.3). Digital camera was used for photography and the images were directly transferred to computer [5].
Southern hybridization
VT1 probe preparation
The amplified DNA fragment of VT1 (348 bp) from E. coli O157:H7 was purified after PCR method using PCR purification kit purchased from Pure-Gene, Malaysia. Pu-rified PCR product was then labeled by Horse Radish Peroxidase using North2South® Direct HRP Labeling and Detection kit (Pierce, USA) to produce VT probe.
Blotting
After electrophoresis on agarose gel, the gel was blotted to positively charged nylon membrane (Hybond N+ membrane, 0.45 μm, Millipore) overnight by capillary transfer. The gel was set up for capillary transfer/alkaline transfer as described in details by Southern [11]. The ny-lon membrane was then dipped in 6X SSC solution and air dried, baked in an oven at 800C for 2 hours.
Hybridization
Prehybridization, hybridization and post hybridization were carried out using North2South® Direct HRP Label-ing, Hybridization and Chemiluminescent Detection kit (Pierce, USA). For detection, the membrane was placed into a film cassette and the blot was exposed to film for 1 min in a dark room and developed by using a developer in University of Malaya Medical Centre, Kuala Lumpur, Malaysia.
Sequencing
The VT1 gene was amplified by PCR method, pooled, purified and sent for sequencing service to BioSyntech Sdn. Bhd., Selangor, Malaysia. The sequencing result was analyzed online using NCBI and BLAST.
Results
PCR
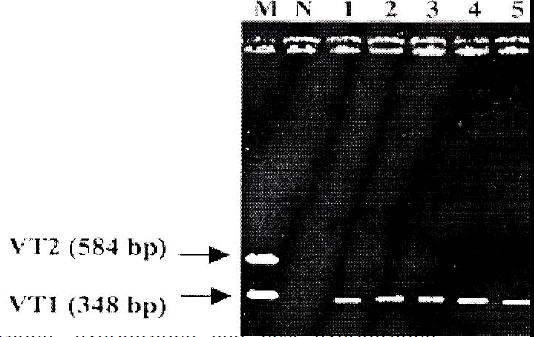
PCR was carried out using all the 30 EPEC isolates. E. coli O157:H7 EDL 933 was used as positive control and sterile deionized water was used as negative control. Ten (33%) out of 30 EPEC isolates showed to carry VT1 gene (348 bp), whereas none of them carried VT2 gene (Fig 1).
DNA extraction
Plasmids and chromosomal DNA were extracted from all the VT1-positive EPEC isolates followed by DNA hy-bridization. All the isolates harbored 1 or more plasmid with the maximum of 4 plasmids.
Hybridization
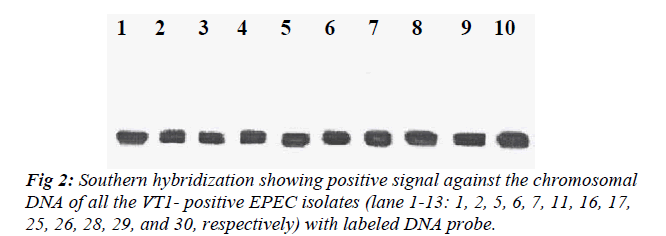
All the VT1-positive EPEC isolates were subjected to Southern hybridization to detect whether the VT1 gene was chromosomally or plasmid mediated. DNA probe showed homology with the chromosome of all the VT1-positive EPEC isolates but none of the plasmid showed homology with the DNA probe (Fig 2).
Sequencing
The amplified VT1 gene (348 bp) from the chromosome of an EPEC strain (E. coli 07) was purified using PCR purification kit purchased from Pure-Gene, Malaysia. The purified amplified DNA was sent for sequencing service to BioSyntech Sdn. Bhd., Selangor, Malaysia. The se-quencing results were analyzed online using NCBI and BLAST software. Both the forward and reverse comple-ment showed 96% homology with the known VT1 gene sequence of VT producing E. coli (O48:H21, O157:H7).
Discussion
The objective of this study was to detect the VT gene in EPEC using PCR protocol and its molecular characteriza-tion. Thirty EPEC isolates were examined for the detec-tion of VT gene. The EPEC isolates were isolated from infants with diarrhea. No other clinical symptoms were observed from the patients. Previously, PCR assays for detection of VT used a single primer pair, which was ca-pable of amplifying both VT1 and VT2 genes [12]. But most methods now include two primer pairs in the same reaction that yield different sized products for VT1 and VT2 [13]. Therefore, in this study two pairs of primer (VT1 and VT2) were used in polymerase chain reaction (PCR) amplification procedure to detect VT1 and VT2 genes. Primer specific amplification was not detected when the DNA template was isolated from VT-negative E. coli, also no false positive amplification was observed from the negative control used, indicating that the method was specific, sensitive and reliable.
Ten EPEC isolates (33%) were found to carry the VT1 gene whereas none of them carried the VT2 gene. These data suggest that VT1 is the most common verotoxin a-mong the EPEC strains isolated from clinical sources in Malaysia. This is in agreement with Nazmul et al. [5] where they showed a similar finding in non-O157 E. coli. VT2 gene was shown to be dominant among E. coli O157 strains isolated from the beef in Malaysia [14]. In another study, VT1 was found to be the most common toxin among the bovine isolates and VT2 the most common in the porcine isolates [15]. Furthermore, Scotland et al. [16] also informed that some strains of EPEC belonging to serogroups O26, O55, O111 or O128 produce VT where they found 18 of the 122 EPEC strains to be VT positive after PCR in United Kingdom. In addition, Scotland et al. [17] have detected VT genes in 57 of 402 (14%) EPEC isolates using PCR method in the United Kingdom. In this study, the occurrence of only VT1 gene may be due to all the isolates have been originated from clinical isolates and/or due to the geographical distribution of the isolates. It may also be possible that the isolates may be the mem-bers of one clone that has become widely distributed.
Plasmids were detected in all VT-positive isolates. These isolates appear to harbor 1 or more plasmids with the maximum of 4 plasmids. VT1 probe showed homology with the chromosomes of all the VT1-positive isolates and suggesting that the VT1 gene to be located on the chromosomes. VT1 gene showed no homology with any of the plasmids. Nazmul et al. [5] and James and James [12] also reported similar findings where they showed that after DNA-DNA hybridization that the VT gene was chromosomally located in all the VT-positive E. coli iso-lates tested. In this study, the VT1 gene from E. coli 07 isolate was amplified and subjected to nucleotide se-quencing. Both the forward and reverse complements showed 96% homology with the known VT1 gene se-quence of VT producing E. coli (O48:H21, O157:H7) and other VTEC isolates. This confirms that the VT1 gene in the present study to be the similar gene carried by other ancestors.
In summary, the PCR method used here was sensitive, specific and reliable. VT1 was found to be the most common verotoxin among the EPEC strains isolated from clinical sources in Malaysia. VT1 gene was located on the chromosome of all the EPECs but how all the VT1-positive EPEC isolates acquired the VT1 gene has yet to be determined. However, studies are in progress to deter-mine the carrier of these VT genes.
References
- Browne MR. Traditional enteropathogenic Escherichia coli of infantile diarrhoea. Rev Infect Dis 1987; 9: 1.
- O?Brien AD, RK Holmes. Shiga and Shiga-like toxins. Microbiol Rev 1987; 51: 206-220.
- Marques LRM, Moore MA, Wells JG, Wachsmuth JK, O?Brien AD. Production of Shiga-like toxin by Es-cherichia coli. J Infect Dis 1986; 154: 338-41.
- Cleary TG, Mathewson JJ, Faris E, Pickering LK. Shiga-like cytotoxin production by enteropathogenic Escherichia coli serogroups. Infect Immun 1985; 47: 335-7.
- Nazmul MHM, Salmah I, Jamal H, Ansary A. Detection and molecular characterization of verotoxin gene in non-O157 diarrheagenic Escherichia coli isolated from Miri hospital, Sarawak, Malaysia. Biomedical Research 2007; 18: 39-43.
- Echeverria P, J Seriwatana, O Sethabutr, A Chatkae-morakot. Detection of diarrheagenic Escherichia coli using nucleotide probes. In: Gene Probes For Bacteria. Academic Press, London. (1990).
- Dorn CR, SM Scotland, HR Smith, GA Willshaw, B Rowe. Properties of verocytotoxin-producing Es-cherichia coli of human and animal origin belonging to serotypes other than O157:H7. Epidemiol Infect 1989; 103: 83-95.
- Pollard DR, WM Johnson, H.Lior, SD Tyler, KR Ro-zee. Rapid and Specific Detection of Verotoxin Genes in Escherichia coli by the Polymerase Chain Reaction. J Clin Microbiol 1990; 28: 540-545.
- Cebula TA, WL Payne, P Feng. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol 1995; 33: 248-250.
- Salmah I. Molecular studies of Pasteurella multocida vetereinary isolates. M.Sc. thesis, University of Malaya, Malaysia, 1997.
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 1975, 98: 503-517.
- Uma MB. Detection and molecular analysis of Shiga toxin producing Escherichia coli isolated from food and clinical source. M.Sc. thesis, University Malaya, Malaysia, 2003.
- James P Nataro, James B Kaper. Diarrheagenic Es-cherichia coli. Clin Microbiol Rev 1998; 11: 142-201.
- Son R, AM Sahilah, G Rusul, A Zainuri, M Tadaaki, A Norio, BK Yung, O Jun, M Nishibuchi. Detection of Escherichia coli O157:H7 in beef marketed in Malaysia. Appl Environ Microbiol 1998; 64: 1153-1156.
- Hales BA, JN Fletcher, G Ridha, RM Batt, CA Hart, JR Saunders. Incidence of common DNA sequences in bo-vine and porcine Escherichia coli strains causing diarrhoea. Res Vet Sci 1991; 50: 355-7.
- Scotland SM, GA Willshaw, HR Smith, B Said, N Stokes, B Rowe. Virulence properties of Escherichia coli strains belonging to serogroups O26, O55, O111 and O128 isolated in the United Kingdom in 1991 from patients with diarrhoea. Epidemiol Infect 1993 111: 429-438.
- Scotland SM, HR Smith, T Cheasty, B Said, GA Will-shaw, N Stokes, B Rowe. Use of gene probes and adhesion tests to characterize Escherichia coli belonging to enteropathogenic serogroups isolated in the United Kingdom. Med Microbiol 1996; 44: 428-443.