ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2019) Volume 30, Issue 2
Molecular detection of Escherichia coli in Wadi Hanifa, Saudi Arabia
1Botany and Microbiology Department, Science College, King Saud University, Riyadh, Saudi Arabia
2Biochemistry Department, Science College, King Saud University, Riyadh, Saudi Arabia
3Laboratory of Plant Biotechnology Applied to Crop Improvement, Faculty of Science, University of Sfax, Tunisia
- *Corresponding Author:
- Nadine Moubayed
Botany and Microbiology
Department Science College King Saud University Saudi Arabia
Accepted Date: January 25, 2019
DOI: 10.35841/biomedicalresearch.30-19-026
Visit for more related articles at Biomedical ResearchBackground: The threat of controlling water borne infectious diseases was always a challenge for health authorities because water related diseases are a major cause of morbidity and mortality worldwide. Faecal coliforms such as Esherichia coli (E. coli) are the indicators most commonly analysed to evaluate the level of faecal contamination in water. Objectives: The study aimed in the first part to compare different techniques for the best detection of E. coli from water. The chosen method was applied in the second part on samples from Wadi Hanifa and neighboring wells. Colonies considered as E. coli were subjected to single colony PCR targeting the genes hha (indicator of enteric bacteria) and tuf (indicator of E. coli), in order to determine if there is a correlation between positive results given with both genes, thus the gene hha could be used as an indicator of fecal contamination in water. Material and methods: 3 techniques of water concentration for the detection of E. coli in artificially contaminated tap water with K12 (E. coli) were compared: (i) The European directive ISO 9308-1 (ii) Ultrafiltration method (UF) and (iii) Membrane filtration followed by direct isolation of genomic DNA. The best technique was applied for E. coli detection in 156 samples collected monthly and during one year from Wadi Hanifa and neighboring wells in Riyadh. Presumptive E. coli strains were subjected to single colony PCR targeting the genes hha and tuf. Results: The comparison of the three techniques has shown that the ISO 9308-1 was more reliable, accurate and economic than the other two tested techniques. Ten (10) E. coli strains (6.41%) were found exclusively in the surface water of Wadi Hanifa whereas; no E. coli strain was detected in wells, fortunately. All E. coli strains tested positive for the gene tuf, were found also positive for the gene hha. Conclusion: The ISO 9308-1 was the technique of choice for the detection and enumeration of coliforms and E. coli in water samples. A good correlation was found between the two genes suggesting that the gene hha could be used as indicator of fecal contamination in water samples.
Keywords
Tuf gene, hha gene, Single colony PCR, Ultrafiltration, Escherichia coli (E. coli).
Introduction
The Kingdom of Saudi Arabia has made comprehensive improvements in all sectors, which have been coupled with high growth rates both in population size and in living standards. This increase in population size and living standards has resulted in an increased demand for water. The majority of required water is supplied either by depleting non-renewable groundwater or by salt water desalination [1]. In Saudi Arabia, surface water sources like manmade well, natural lakes and open water reservoirs are almost exploited due to every conceivable use. Typically, the wastewater that is discharged from most of the Saudi wastewater stations is the water that has not reached the secured stages 3-4, and therefore, this wastewater pollutes the environment, especially all water sources near metropolitan areas. This contamination with wastewater has made surface water resources in Saudi Arabia highly polluted, especially in parched valleys [2]. Uncontrolled industrial wastewaters result, on the other hand, in contamination of surface water and consequently an imbalance of ecosystem [3,4]. In developing world, 70% of untreated industrial wastewaters are released into surface water [5]. Hence surface water quality control must be strongly determined to reduce the public health risks. This quality control however, relies mainly on detecting and enumerating the presence of coliforms and E. coli. It was estimated that 80% of communicable diseases in the world are waterborne [6], thus analysing the presence of total coliform and E. coli in water samples is the most commonly used method to test the hygienic quality of water samples [7,8].
The bacterial coliform group has been used extensively as an indicator of water quality analysis and has historically led to the public health protection concept. The use of the coliform group as an indicator of fecal contamination is subject to strict governmental regulations. E. coli is the most common coliform among the intestinal flora of warm-blooded animals and its presence might be principally associated with fecal contamination, hence no E. coli are therefore allowed in drinking water [9]. In order to protect the human health, there is a need to control the quality of waters. Chemical, physical and biological parameters are used for the water characterization. Biological characterization is done by means of the coliforms enumeration and other bacteria. The most commonly tests used in the water industry are the identification and detection of coliforms and E. coli [10]. Classic microbiological indicators such as faecal coliforms, E. coli and Enterococci are the 2 indicators most commonly analysed to evaluate the level of faecal contamination in water [11-13].
Microbial analysis is particularly very important with regard to recycling of wastewater treatment and its reuse. Different microbial pathogens have different infectious doses. Thus, the qualitative and quantitative detection of different microbial pathogens in the water and wastewater samples is imperative. An ideal detection method should be fast, sensitive, highly accurate, and easy to perform and also inexpensive. There are a number of established methods for the detection of most microbial pathogens in water and wastewater. The methodology specified in the European Directive for the detection of E. coli is the method described by ISO (ISO 9308-1) and it is widely used. Molecular identification of water contaminants mainly E. coli has also been extensively used, in the last decades, particularly the polymerase chain reaction (PCR). This molecular technique allows very specific and rapid detection with high degree of sensitivity and specificity depending; however, on the choice of the primers sets [14,15].
The aim of our study was to examine the methods currently in use which can be proposed for the monitoring of coliforms and E. coli in water which defines the coliforms. We have compared 3 techniques of water concentration for the detection of E. coli in artificially contaminated tap water with the strain E. coli K12. (i) The European directive ISO 9308-1 was applied to examine and monitor coliforms and E. coli in water. (ii) Ultrafiltration method (UF), based on the use of Amicon Ultra-15 Centrifugal filter which is an Ultracel regenerated cellulose membrane with high capacity to retain microorganisms, nucleic acids and finally (iii) Membrane filtration followed by direct isolation of genomic DNA from membrane filtered water samples using the “Rapid Water DNA isolation kit”. The best technique was applied on 156 surface water samples of Wadi Hanifa, Riyadh and neighboring wells collected from January 2015 to December 2015, for the detection of E. coli. Colonies considered presumptively as E. coli were subjected to single colony PCR targeting the genes hha and tuf, in order to determine if there is a correlation between positive results given with the hha gene (indicator of enteric bacteria) and the gene tuf (indicator of E. coli).
Material and Methods
Water samples
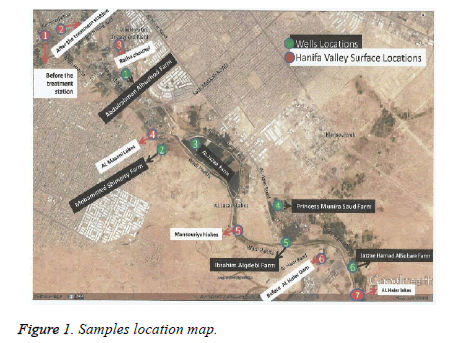
156 water samples were collected from surface water of Wadi Hanifa and neighboring wells, during the period of January 2015 to December 2015. Surface water samples were collected from 7 different locations of Wadi Hanifa, (Figure 1) and designated from S1-S7. Similarly, well water samples were collected from 6 different locations (W1-W6). The collected water samples in sterile tubes of 100 ml were directly transported at 4℃ to the laboratory for the microbiological assays.
Bacterial strain
The bacteria used in the present study was K12 (E. coli MG1655) which was gently offered by Antonio Juarez, professor of microbiology, group leader at the institute for bioengineering of Catalonia (IBEC), Barcelona, Spain.
Artificial contamination of tap water by E. coli
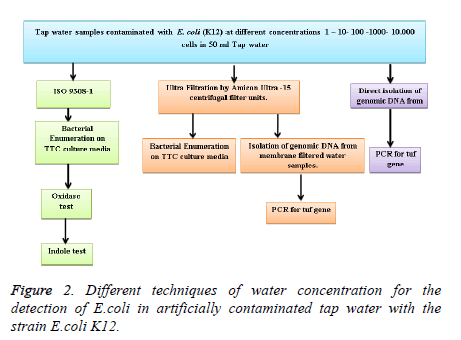
The strain K12 (MG 1655) was cultured overnight on nutrient broth to reach a concentration between 2 × 108/ml and 2 × 109/ml. Then, serial dilution was performed and 1 ml from 10 ml overnight culture was taken aseptically and added to 9 ml sterile Ringer solution. 100 μl of the last 2 dilutions (10-6, 10-7) were plated on top of Agar plates at 37°C for 18-24 hours for direct cell count and hence to be used for the sample water artificial contamination. In order to assess the sensitivity of our assay, water samples were contaminated with different cell concentrations, depending on the overnight cell concentration estimated and recorded. Contaminated water samples ranging from 1 to 104 cells/ml were used to check the sensitivity of the assay. Three techniques of water concentration were compared for the detection of E. coli in artificially contaminated tap water with K12 (Figure 2).
ISO 9308-1(Membrane filtration method)
The ISO 9308-1 describes a reference method (Standard test) for the detection and enumeration of E. coli and coliforms bacteria. The method is based on membrane filtration, subsequent culture on a chromogenic coliform agar medium, and calculation of the number of target organisms in the water sample. The method consists of two parts: 1/ the water samples were filtered through a cellulose esters membrane of 47 mm of diameter (0.45 μm filters, millipore). The filter obtained was then placed on special coliforms media Tergitol 7 Agar (TTC), and incubated for 24 hours at 37℃ [16] (Bartram and Pedley, 1996). 2/ Biochemical characterization: oxidase and indole tests of the typical lactose positive colonies were done leading to the detection and enumeration of E. coli.
Ultrafiltration method
Ultrafiltration is a kind of membrane filtration in which forces like pressure or concentration gradients lead to a separation through a semipermeable membrane. In our study, the UF was based on the use of Amicon Ultra-15 Centrifugal filter which is an Ultracel regenerated cellulose membrane with high capacity to retain microorganisms, nucleic acids and enzymes. 15 ml of sample water was added to Amicon Ultra filter-tube then placed into centrifuge rotor, and centrifuged at 4,000 × g and 4℃ for approximately 15 minutes (Heraeus Megafuge 16 R Centrifuge, Thermo Scientific). The concentrated water sample (about 200 μl) was then used for cultural analysis.
Direct isolation of genomic DNA from membrane filtered water samples
The RapidWater® DNA isolation kit (MOBIO) was used for the isolation of bacterial genomic DNA from membrane filtered water samples. The Power Water® DNA Isolation Kit starts with the filtration of a water sample onto a filter membrane (0.45 μm, Millipore). The membrane is then added to a special 5 ml bead beating tube containing a unique bead mix. Rapid and thorough lysis occurs through vortex mixing in a reformulated lysis buffer that enhances the isolation of microorganisms from filter membranes. After the protein and inhibitor removal steps, total genomic DNA is captured on the silica spin column. High quality DNA is then washed and eluted from the spin column membrane for the use of PCR.
Polymerase chain reaction (PCR)
PCR was done targeting two genes present in the genomic DNA of E. coli: tuf and hha genes. The reaction mixture is the same for each with the exception of primers and PCR program. The PCR reagent kit (HotStarTaq Master Mix, Qiagen) was used to amplify the region of interest in all DNA samples. Two couples of primers were used for PCR amplification for the genes hha and tuf. The primers specific for the gene tuf were: TEcol553/TEcol754 which amplify a region of 220 bp. The primers specific for the gene hha were: hhaFW/hhaRV, which amplify a region of 166 bp [17]. The reactions were performed in a final volume of 20 μl. The DNA undergoes a polymerization reaction in a thermal cycler (T100 PCR Thermal Cycler, BIO-RAD) using the same program for the two genes except for the annealing temperature for the primers. The program consists of: initial denaturation at 95℃ for 5 minutes and 35 cycles at 95℃ for 30 sec; annealing at 55℃ for 30 sec (for hha gene) or at 58℃ for 30 sec (for tuf gene) and, primer extension at 72℃ for 30 sec followed by a final extension at 72℃ for 10 min.
Results
Set up of the water concentration techniques for the best recuperation of E. coli
The European directive ISO 9308-1 was applied to examine E. coli in artificially contaminated tap water with different bacterial concentration (1-104 cell/ml). The results obtained were summarized in the Table 1 below. The number of bacterial colonies in the membrane filtration technique was ranging between 35-300 colonies which indicate a very good correlation between the number of E. coli seeded and the number of E. coli recovered after the application of the ISO 9308-1.
| Bacterial concentration (cell/mL) |
ISO 9308-1 | Ultrafiltration |
|---|---|---|
| CFU | CFU | |
| 1 | 35 | 20 |
| 10 | 140 | 13 |
| 100 | 186 | 18 |
| 1000 | 280 | 21 |
| 10 | 300 | 26 |
Table 1: E.coli colonies enumeration after the application of the ISO 9308-1.
For the ultrafiltration technique, 15 ml of the tap water sample contaminated with different bacterial concentration was added to Amicon Ultra filter-tube then centrifuged. The extract was recuperated then placed into a filter membrane. Two assays were followed at the same time: (1) the filter was incubated in TTC agar plates for coliforms enumeration, before the biochemical characterization was done. (2) The filter was used to direct isolation of genomic DNA with the “Rapid Water DNA isolation kit”. Then a PCR was done with specific primers for the detection of tuf gene (E. coli indicator) to check the sensitivity of the technique of ultrafiltration. The results obtained after E. coli colonies enumerations were summarized in the Table 1. The number of bacterial colonies was ranging between 13-26 colonies which is very low and does not reflect the real number of bacteria seeded. The reduced number of bacteria in the ultrafiltration technique may be due to the rupture of the bacterial cell wall by a high centrifugal speed or by the adhesion of bacteria on the wall of the tube and this may affect the number of colonies developing. On the other hand, the results obtained after genomic DNA extraction followed by DNA quantification and PCR with specific primers for the detection of tuf gene, are shown in Table 2. This comparative study aimed to select the best assay that give better results even with very low amount of E. coli concentration, such as 1 cell/ml. Surprisingly, very low amounts of DNA were obtained (between 6.8 ng/μl and 13 ng/μl). Due to the low quantity of DNA obtained, PCR analysis gave negative results.
| Bacterial concentration (cell/mL) |
DNA (ng /µL) After Ultrafiltration |
DNA (ng /µL) After membrane filtration |
|---|---|---|
| 1 | 6.8 | 10.3 |
| 10 | 4.2 | 10 |
| 100 | 11.9 | 12.5 |
| 1000 | 13 | 13.9 |
| 10 | 11.6 | 13.8 |
Table 2: DNA quantification (ng /µl) followed by direct isolation of genomic DNA after Ultrafiltration and membrane filtration techniques.
Finally, we have detected E. coli by Membrane filtration followed by direct genomic DNA isolation. The RapidWater® DNA isolation kit (MO BIO) was used for the isolation of bacterial genomic DNA from membrane filtered water samples (100 mL). Table 2 below summarizes the results obtained after DNA quantification of total DNA extracted from the filter membrane. As we have seen in the previous experiment with ultrafiltration, very low amounts of DNA were obtained (between 10 ng/μl and 13.9 ng/μl). The low quantity of DNA obtained, may explain why PCR analysis with specific primers for tuf gene gave negative results. Data obtained from the comparison of the three techniques have shown that the ISO 9308-1 was more reliable, accurate and economic than the two others tested techniques.
Application of the ISO 9308-1 for the detection of E. coli and coliforms in surface and wells water samples of Wadi Hanifa
Detection of coliforms and E. coli by the ISO 9308-1 protocol: 100 ml of each surface water sample (diluted 100X) and well waters (not diluted) were filtered. Membrane filters were then loaded on TTC and MAC media and incubated at 37℃ for 24 hours for coliforms detection. Colony count was determined as an average of duplicated experiments. All the counted bacterial colonies that grew on TTC are colored in red, orange, yellow and green, while the colonies grown in MAC media were colored in red.
For surface water, we have observed that there was a higher bacterial number in TTC agar plates (300 CFU) in surface water samples mainly at location S6 accordingly to the months of: January, February, March, May, August, September and December; and the location S3 in February and September. A low bacterial number was detected in the location S5 in April (67 CFU). The higher bacterial number observed with MAC plates was 30 CFU in S3 (June). The lowest number (1 CFU) was detected in S2, S3, S4, S5 and S6 during the months of: June, July, August, September, October, November and December.
With regard to well water samples, we have detected a higher bacterial number in TTC agar plates (140 CFU) in W6 in March. The lowest number was observed at location W5 in January. For MAC, the number of colonies was much lower and ranging between 0 and 11 CFU/ml, in the location W6/ August and W3/June, respectively.
As described by the European directive ISO 9308-1, biochemical and cultural steps are mandatory for the identification and enumeration of E. coli. 50 colonies were identified using oxidase test from surface water samples and 36 colonies were found for well samples. The indole test was made for the isolated 86 colonies positive for oxidase test. The results obtained indicated the presence of 10 colonies of E. coli, in surface water samples. No detection of E. coli in well water samples. For further confirmation of the results obtained with oxidase and indole tests, which gave as a result 10 E. coli colonies, another confirmatory test was used, which is the Eosin Methylene Blue Agar (EMB). The obtained results are completely in accordance with the two previous tests (oxidase and indole tests) which have indicated that only 10 colonies of E. coli could be detected among 156 samples collected from surface water of Wadi Hanifa and neighboring wells.
Relationship between tuf and hha genes in the detection of E. coli colonies
The 10 colonies considered presumptively as E. coli were subjected to single colony PCR targeting the genes hha and tuf. Our objective was to determine if there is a correlation between positive results given with the hha gene (indicator of enteric bacteria) and the gene tuf (indicator of E. coli). A good correlation will suggest that hha gene could be used as an indicator of the presence of fecal contamination in water. Each E. coli colony was mixed with 50 μl of purified water in the first step, and then 2 μl were added directly to two PCR reactions: one reaction with specific primers for tuf gene (TEcol553/TEcol754) and another one reaction for hha gene (hhaFW/hhaRV). The results showed the amplification of the tuf gene for all E. coli colonies, giving a PCR product of 220/258 bp in size. For the gene hha, 8 colonies from 10 colonies could be amplified, giving a PCR product of 166 bp in size.
Table 3 showed the distribution of the 10 colonies depending on the months and the locations. We can see that the two missing colonies were from the same location: S3 and for two consecutive months: July and August. Since all E. coli strains tested positive for the gene tuf, were found also positive for the gene hha, except two strains, we can say that there is a good correlation between the two genes suggesting that the gene hha could be used as indicator of fecal contamination.
| Month | Sample | Gene tuf | Gene hha |
|---|---|---|---|
| March | 7S | + | + |
| April | 1S | + | + |
| June | 2S | + | + |
| 1S | + | + | |
| July | 2S | + | + |
| 3S | + | - | |
| August | 3S | + | - |
| September | 1S | + | + |
| October | 1S | + | + |
| December | 1S | + | + |
Table 3: The PCR results of surface water samples.
Discussion
Data obtained from the tap water experiments revealed that the best method for water samples concentration was the European directive (ISO 9308-1). It is more reliable than the ultrafiltration and bacterial genomic DNA isolation methods, easy to handle and more economic. The ISO 9308-1 was the technique of choice for us for the subsequent detection and enumeration of coliforms and E. coli in real samples of water, in the second part of the study. Our results are in accordance with those reported by Paradis et al. [18] and Afnor [19] who reported that the membrane filtration technique is the method most widely used for the enumeration of coliforms in drinking water in Europe [20], have also demonstrated that PCR methods offer a higher level of specificity detection. Nevertheless, they have major limitations when applied to natural samples, including low amplification rates linked to the presence of inhibitor substances and the lack of information on the physiological activity of cells.
Bacterial enumeration of coliforms and E. coli in surface water samples have observed that there was a higher bacterial number in TTC agar plates (300 CFU) in surface water samples mainly at location S6 accordingly to the months of: January, February, March, May, August, September and December; and the location S3 in February and September. A low bacterial number was detected in the location S5 in April (67 CFU). The higher bacterial number observed with MAC plates was (30 CFU) in S3 (June). The lowest number (1 CFU) was detected in S2, S3, S4, S5 and S6 during the months of: June, July, August, September, October, November and December. Our results are in line with those of Kumar et al. [21] in India, who found that the range of coliforms in Shivnath River was comprised between 80 and 300 CFU. In Riyadh, the lack of drainage network and the use of treated sewage water for irrigation may explain the high microbial load in the surface water of Wadi Hanifa.
Bacterial enumeration of coliforms and E. coli in well water samples was higher on TTC agar plates (140 CFU) in W6 in March. The lowest number was observed at location W5 in January. For MacConky agar plates, the number of colonies was much lower and ranging between 0 and 11 CFU/ml, at location W6/August and W3/June, respectively. These results were in line with those of Kumar et al. [21] in India and Todorov et al. [22] in Bulgaria, who found that the range of coliforms in well water samples was comprised between 10 and 140 CFU.
All E. coli strains tested positive for the gene tuf, were found also positive for the gene hha, except two strains. Since all E. coli strains tested positive for the gene tuf, were found also positive for the gene hha, except two strains, we can say that there is a good correlation between the two genes suggesting that the gene hha could be used as indicator of fecal contamination. The ease and effectiveness of colony polymerase chain reaction (PCR) has allowed rapid amplification of DNA fragments and screening of large number of colonies. This colony PCR technique can replace the fastidious process of isolating genomic DNA and make possible the rapid amplification of genomic DNA and largescale screening.
Conclusion
The present study emphasized the use of the ISO 9308-1 as the technique of choice for the determination of the water quality control when compared to other techniques applied in this study naming Ultrafiltration and Membrane filtration followed by direct isolation of genomic DNA. Moreover, the molecular analysis of E. coli using both the tuf and hha gene suggested that the gene hha could be used as indicator of fecal contamination.
Acknowledgment
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award number (12-ENV2528-02).
References
- Directorate of Census Operations (2011) District census handbook, Imphal East, Village and Town Directory, Series-15, Part-XII-A p: 166.
- Directorate of Census Operations (2011) District census handbook, Thoubal, Village and Town Directory, Series-15, Part-XII-A p: 133.
- Central Ground Water Board (2013) Ground water information booklet, Imphal East District, Manipur. Technical Report Series: D No: 07/2013-14 p: 23.
- Central Ground Water Board (2013) Ground water information booklet, Thoubal District, Manipur. Technical Report Series No: 27/2013-14 p: 15.
- Roy PS, Saran S, Ghosh S, Prasad N, Talukdar G (2002) Development of biodiversity information system for North East India using Internet GIS. International Archives of Photogrammetry Remote Sensing and Spatial Information Sciences 34: 361-370.
- SAC (2007) Nationwide forest encroachment mapping using remote sensing and GIS techniques-Manipur State. Jointly carried out by Space Applications Centre (SAC-ISRO), Ahmedabad, Manipur Forest Department (MFD), Imphal, Manipur State Remote Sensing Applications Centre (MARSAC), Imphal, Technical Report p: 92.
- Khan A, Ahmad S, Khurshid S (2012) Geology and geomorphology of the Manipur Valley using digitally enhanced satellite image and SRTM DEM in the Eastern Himalaya, India. Int J Geosci 3: 1010-1018.
- Planning Department, Government of Manipur (2014)New land use policy/project of Manipur. An Approach Paper on New Land Use Policy/Project, Manipur p: 32.
- Singh AM, Devi KR (2016) Land use and land cover change detection of fringe areas of Imphal City, Manipur, India. IOSR J Humanit Soc Sci 21: 9-16.
- Mruthyunjaya RK, Raghu V, Samdup T, Rajesh V (2015) A web-based application study for integrated land management and administrative planning for East and South Districts of Sikkim State, India. J Geomatics 9: 146-152.
- National Remote Sensing Agency (2006) National land use land cover mapping using multi-temporal satellite data. Manual p: 125.