ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 13
Molecular docking and in silico ADMET study reveals 3-(5-{[4-(aminomethyl) piperidin-1-yl] methyl}-1h-indol-2-yl)-1h-indazole-6-carbonitrile as a potential inhibitor of cancer Osaka thyroid kinase
Afzal Hussain* and Chandan Kumar Verma
Department of Bioinformatics, MANIT, Bhopal, Madhya Pradesh, India
Accepted on May 17, 2017
COT (Tpl2/MAP3K8) is a serine/threonine kinase that plays a major role in the TNF-alpha production through MEK, ERK pathway and the production of other pro-inflammatory cytokines such as IL-1 beta. It's crucial role in inflammatory diseases and cancer, for finding out the best potent anticancer agent glide tool of the Schrodinger software was used for virtual screening using ligand databases like Drug Bank, MDPI and MayBridge Hitfinder against the target protein. The best compound was identified which can bind in the active site fully and showing good docking score as well as good pharmacokinetics characteristics. ADMET, MM-GBSA and DFT analysis was also done on the selected screened compounds. The compound 3-(5-{[4-(aminomethyl) piperidin-1-yl] methyl}-1H-indol-2-yl)-1Hindazole- 6-carbonitrile (DB07075) was found to be most potent among all the screened compounds. Hopefully compound (DB07075) could be used as anti-cancer agent against cancer, rheumatoid arthritis and various death leading diseases.
Keywords
COT kinase, Drug bank, Cancer, MDPI, Inhibitors, Potent.
Abbreviations
SBVS: Structure Based Virtual Screening; COT: Cancer Osaka Thyroid Kinase; DFT: Density Functional Theory; MW: Molecular Weight; HOMO: High Occupied Molecular Orbital; LUMO: Lowest Un-Occupied Molecular Orbital; MDPI: Molecular Diversity Preservation International; MH: MayBridge HitFinder; BBB: Blood Brain Barrier; HIA: Human Intestinal Absorption.
Introduction
Protein kinases play a very vital role in the cell biology where it can modify the functioning of a protein in the cell. Protein phosphorylation is a process which can increase or decrease the enzyme activity to alter the cellular processes like transcription and translation and play a major role in signal transduction pathways which can activate other protein to be functioned. Dysregulation of the protein kinases is related with many diseases such as cancer and inflammatory disorders [1,2].
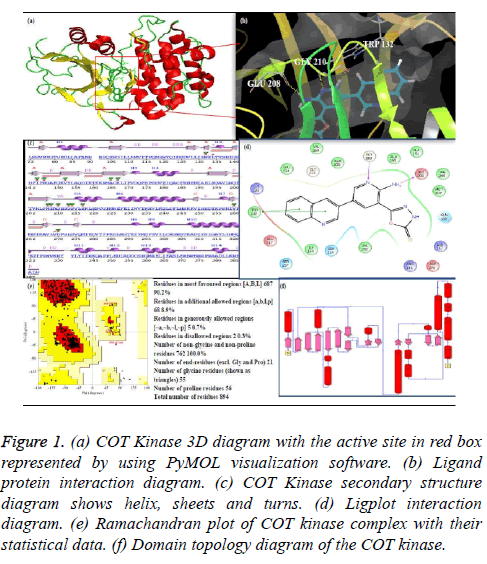
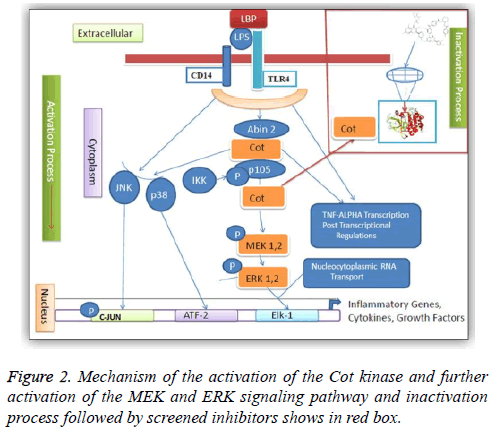
One of the protein kinase is Cancer Osaka Thyroid Kinase which is a Mitogen Activated Protein Kinase 8 (MAP3K8) (Figure 1). It involve to regulate various signaling pathways like cell survival, cell proliferation and cell death as well as various inflammatory pathways including ERK, JNK, p38, NF- κB and plays an important role in the immune system also. COT kinase activation has been done with various proinflammatory stimuli like Lipopolysaccharide (LPS), TNF, CD40 ligand through Toll-Like Receptors (TLRs), TNF receptor 1, CD40 and Interleukin (IL-1) receptor [3]. COT kinase plays an important role for the activation of the downstream pathway like MEK/ERK with the stimulation of the pro-inflammatory receptors IL-1/TNF/TLR, belongs to the receptor super families, while MAP3K Tak1 facilitate to activate the p38, JNK and NF-kB pathways (Figure 2). Talking about the structure configuration, COT kinase forms a heterotrimeric complex together with NF-kB1 precursor protein p105 and the ubiquitin-binding protein ABIN-2. Its activation depends on the IKKB which triggers proteasomal degradation of p105 and releases the ABIN-2 from the association with p105 and COT [4-6].
Figure 1: (a) COT Kinase 3D diagram with the active site in red box represented by using PyMOL visualization software. (b) Ligand protein interaction diagram. (c) COT Kinase secondary structure diagram shows helix, sheets and turns. (d) Ligplot interaction diagram. (e) Ramachandran plot of COT kinase complex with their statistical data. (f) Domain topology diagram of the COT kinase.
COT kinase involvement in various diseases like papillary thyroid cancer [7], breast cancer [8-11], ovarian cancer [12], multiple sclerosis [13], obesity-induced adipose tissue inflammation [14], liver injury [15], metastasis of clear cell renal cell carcinoma [16], androgen depletion-independent prostate cancer growth [17], obesity [18], pancreatic and lung inflammations [19].
In the literature study we have found the inhibitors like indazoles [20], thieno (2, 3-c) pyridine [21,22], 4-alkylamino- (1, 7) naphthyridine-3-carbonitriles [23], 8-substituted-4- anilino-6-aminoquinoline-3-carbonitriles [24], 1, 7- naphthyridine-3-carbonitriles [25], substituted indolyl indazoles [26].
Nowadays the challenge is to find out the small molecule cancer drugs [27,28]. Structure based drug design is the accurate approach for the development of cell cycle inhibitor for cancer therapy. SBVS technique is very important to find out the drug compound very rapidly from the large library of the compounds in the medicinal chemistry research [29]. COT is the most important therapeutic target for cancer research [30]. A lot of inhibitors have been approved which shows inhibition of cancer progression. Therefore, it is a most important work to find out a selective and novel inhibitor, which can recognize the target active site easily for reducing its infectious properties. In this current study the central aim is to find out a selective and most potent inhibitor for reducing the death leading diseases like cancer, cardiac hypertrophy and Alzheimer’s.
Materials and Methods
Retrieval and preparation of the protein structure of COT kinase
The 3D structure of the COT kinase as receptor (PDB ID: 4Y83) was retrieved from the protein data bank (http:// www.rcsb.org/pdb/home/home.do). Protein preparation wizard of the Schrodinger software has been used for preparing the protein structure where bond orders were assigned and hydrogen atoms were added as well as restrained minimization step has also been done using OPLS force field with RMSD cut off of 0.30 A0.
Ligand preparation
Total 62,801 small molecules of the ligand were retrieved from Drug Bank database (http://www.drugbank.ca/), MayBridge HitFinder database (http://www.maybridge.com/portal/ alias__Rainbow/lang__en/tabID__229/DesktopDefault.aspx, and MDPI database (http://www.mdpi.org/cumbase.htm). LigPrep module of the Schrodinger software was used for preparing all the retrieved Ligands where bond order and the bond angle were assigned as well as minimization was done by using OPLS-2005 force field. The Epik option was used for keeping ligand in the correct protonation state [31].
Grid preparation
The generation of the Grid of the receptor protein was done using the Glide protocol of the Schrodinger software where the reference ligand binding site was selected as centroid for Grid where on the active site the screened ligand bind. Scaling factor was selected as 1.0 and the partial charge cut-off was selected as the 0.25 [32].
Preparation of the reference compounds
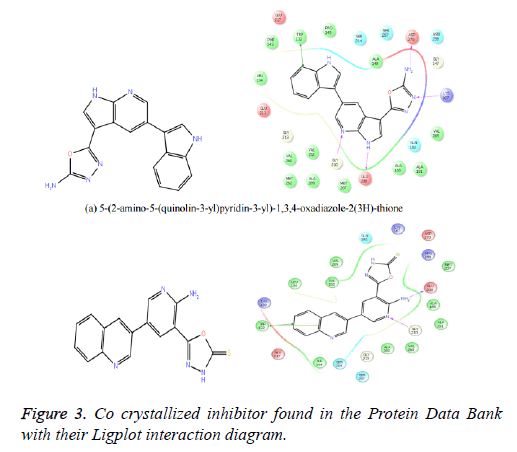
The reference ligand compounds were retrieved from the cocrystallized structure of the protein-ligand structure complex (Figure 3), which is available in the protein data bank (PDB Id: 4Y83) (http://www.rcsb.org/pdb/explore/explore.do? structureId=4Y83). The reference compounds were prepared using a LigPrep module of the Schrodinger software and docked with the prepared receptor structure. This docking score of the reference compounds kept for comparing with the best potent compounds of the Drug Bank, MDPI and MayBridge HitFinder database.
Virtual screening
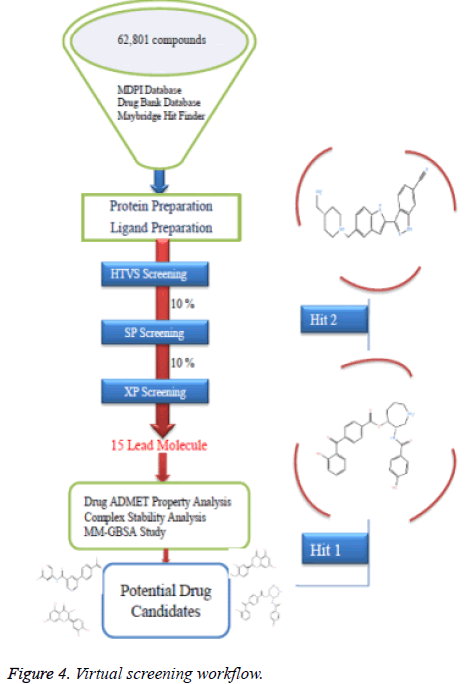
High throughput virtual screening is a bioinformatics technique where we can screen a huge number of ligands against the target protein rapidly. A total number of 62,801 ligand compounds were retrieved from the Drug Bank, MDPI and MayBridge HitFinder databases. The reference compounds were also incorporated in the ligand database. These compounds and the reference compounds were subjected to the Lipinski filtration and reactive functionality. The Glide maestro protocol of the Schrödinger software was used for the virtual screening. Docking was performed in the three different phases like HTVS (High Throughput Virtual Screening), SP (Standard-Precision), XP (Extra-Precision). The library contains a large number of compounds so that after performing HTVS screening remaining 10% was performed using the Standard-precision docking, and remaining 10% was performed using the Extra-precision docking for getting the best potent compounds. The virtual screening workflow is shown in (Figure 4).
Density functional theory analysis
It is also very essential to find out the molecular orbital energy of the selected ligand. For this study HOMO, LUMO molecular orbital energy analysis was done. The gap energy is defined as difference between HOMO and LUMO molecular orbital energy, that energy indicates the excitation energy as well as showing the stability and reactivity of the compounds [33].
Drug like properties analysis
The best potent compounds were selected for finding the drug like properties. These properties follow the Lipinski’s rule of five. The properties which have been considered such as Molecular Weight (MW), Hydrogen Bond Acceptor (HBA), Lipophilicity (log P), Hydrogen Bond Donor (HBD), human oral absorption [34].
ADMET profiles analysis
ADMET properties of the compounds are related to absorption, distribution, metabolism, excretion and toxicity through the human body. The ADMET profile is very essential for evaluating the pharmacodynamic activities of the ligand compounds. In this study we have used the bioinformatic tool admetSAR (http://lmmd.ecust.edu.cn:8000/) [35].
MM-GBSA approach for drug-target binding energy estimation
Molecular mechanics generalized born surface area is used to finding the drug-target binding energy and it was calculated by using Schrodinger software. This binding energy estimates the stability of the protein with the ligand complexes [36].
Result and Discussion
Database screening and docking against the target
Total eleven potent novel lead compounds were found from different databases like Drug Bank, MDPI and MayBridge HitFinder using virtual screening and docking approach against the target protein, which showed best docking score.
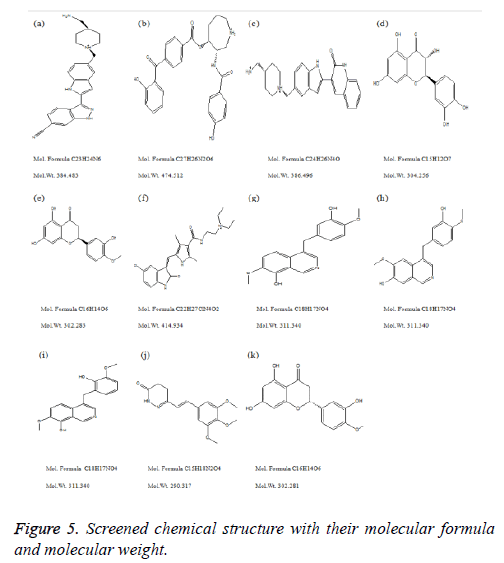
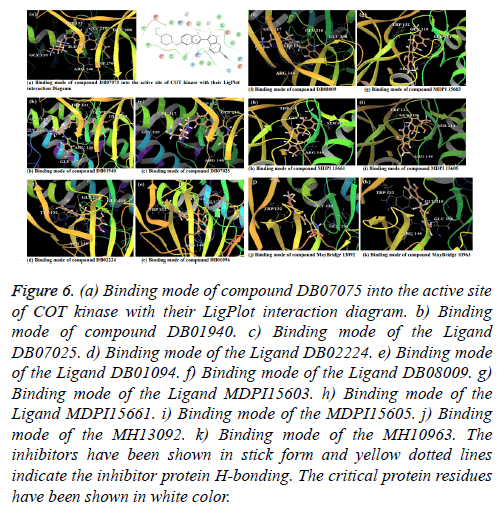
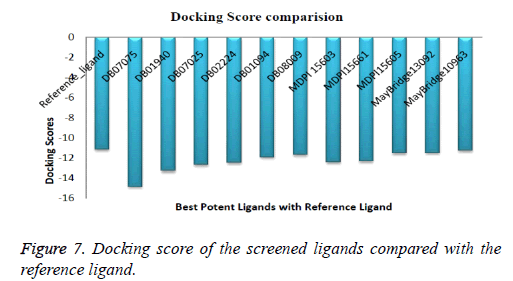
Compound DB07075 was found to be more potent and selective, which interacts with the target protein active site residues. It makes H-bonding with residues GLY 210, GLU 208, ASP 270, and GLU 220, Pi-Pi stacking with TRP 132, ARG 146 and hydrophobic interaction with LEU 134, LEU 216, PRO 145, ILE 144, VAL 269, VAL 152, ALA 209, ALA 165, ALA 191 with docking score -14.860. The docking score is high and it covered the reference ligand binding site completely. It means this lead compound tightly docked in the active site of the target protein and showing good inhibition property. The docking score for DB07075, DB01940, DB07025, DB02224, DB01094, DB08009, MDPI 15603, MDPI 15661, MDPI 15605, MayBridge 13092 and MayBridge 10963 has been identified (Table 1). The docking and ligPlot interaction diagram gives a better view of interaction between target and lead compounds (Figures 5 and 6). The docking score comparisons graph also shown the better comparisons (Figure 7).
| Compound | Compound structure | Mol. Wt. | Mol. formula | Docking score |
|---|---|---|---|---|
| Hit 1 DB07075 |  |
384.483 | C23H24N6 | -14.86 |
| Hit 2 DB01940 |  |
474.512 | C27H26N2O6 | -13.304 |
| Hit 3 DB07025 |  |
386.496 | C24H26N4O | -12.673 |
| Hit 4 DB02224 |  |
304.256 | C15H12O7 | -12.49 |
| Hit 5 DB01094 |  |
302.283 | C16H14O6 | -11.92 |
| Hit 6 DB08009 |  |
414.934 | C22H27ClN4O2 | -11.685 |
| Hit 7 MDPI 15603 |  |
311.34 | C18H17NO4 | -12.408 |
| Hit 8 MDPI 15661 |  |
311.34 | C18H17NO4 | -12.335 |
| Hit 9 MDPI 15605 |  |
311.34 | C18H17NO4 | -11.525 |
| Hit 10 MH 13092 |  |
290.317 | C15H18N2O4 | -11.491 |
| Hit 11 MH 10963 |  |
302.281 | C16H14O6 | |
| 5-(2-amino-5-(quinolin-3-yl)pyridin-3-yl)-1,3,4-oxadiazole-2(3H)-thioner | - | - | - | -11.1403 |
| r=reference compound. | ||||
Table 1. 2D structure of the selected inhibitors from DrugBank, MDPI and MayBridge HItFinder databases and reference compounds respectively with their docking scores.
Figure 6: (a) Binding mode of compound DB07075 into the active site of COT kinase with their LigPlot interaction diagram. b) Binding mode of compound DB01940. c) Binding mode of the Ligand DB07025. d) Binding mode of the Ligand DB02224. e) Binding mode of the Ligand DB01094. f) Binding mode of the Ligand DB08009. g) Binding mode of the Ligand MDPI15603. h) Binding mode of the Ligand MDPI15661. i) Binding mode of the MDPI15605. j) Binding mode of the MH13092. k) Binding mode of the MH10963. The inhibitors have been shown in stick form and yellow dotted lines indicate the inhibitor protein H-bonding. The critical protein residues have been shown in white color.
The series, according to decreasing docking scores is as follows:
DB07075>DB01940>DB07025>DB02224>MDPI15603>MD PI15661>DB01094>DB08009>MDPI15605>MayBridge1309 2>MayBridge10963>Reference_ligand
HOMO LUMO stability analysis of the screened compounds
HOMO LUMO stability analysis was done on selected best potent ligand compounds which show drug and protein interaction stability and contribution of selected inhibitors (Table 2). All the selected ligand showed minimal HOMOLUMO gap with the average energy difference of 0.14 eV, signifying molecular reactivity. The orbital energy and the difference between HOMO and LUMO energy (gap energy) were estimated. HOMO LUMO energy of the inhibitor DB07075 (-0.35210, -0.26869) and remaining inhibitor DB01940 (-0.27794, -0.15153), DB07025 (-0.34105, -0.27646), DB02224 (-0.21787, -0.05554), DB01094 (-0.20716, -0.05019), DB08009 (-0.26440, -0.13994), MDPI 15603 (-0.20165, -0.05099), MDPI 15661 (-0.19274,-0.04465), MDPI 15605 (-0.19934, -0.05680), MH 13092 (-0.19795, -0.05815), MH 10963 (-0.21486, -0.04716). The lowest energy gap was found for DB07075 (0.08341) and DB07025 (0.06459). HOMO energy is higher compared to LUMO energy for all ligand which represent an ability to donate the electrons rather than accept electrons with their partner receptor binding site region.
| S. no. | Compound ID | HOMO energy | LUMO energy | HLG |
|---|---|---|---|---|
| 1 | DB07075 | -0.3521 | -0.26869 | 0.08341 |
| 2 | DB01940 | -0.27794 | -0.15153 | 0.12641 |
| 3 | DB07025 | -0.34105 | -0.27646 | 0.06459 |
| 4 | DB02224 | -0.21787 | -0.05554 | 0.16233 |
| 5 | DB01094 | -0.20716 | -0.05019 | 0.15697 |
| 6 | DB08009 | -0.2644 | -0.13994 | 0.12446 |
| 7 | MDPI 15603 | -0.20165 | -0.05099 | 0.15066 |
| 8 | MDPI 15661 | -0.19274 | -0.04465 | 0.14809 |
| 9 | MDPI 15605 | -0.19934 | -0.0568 | 0.25614 |
| 10 | MH 13092 | -0.19795 | -0.05815 | 0.1398 |
| 11 | MH 10963 | -0.21486 | -0.04716 | 0.1677 |
Table 2. Orbital energy of the selected inhibitors of the DrugBank, MDPI and MayBridge HItFinder databases.
Drug-likeliness property analysis against screened compounds
These selected ligand compounds were evaluated for the drug likeliness properties. These selected hits followed Lipinski’s rule of five. Evidence for drug like characteristics. A good drug compound is recognized which shows effective metabolism, action and absorbed in record time and well distributed throughout the system. The QikProp tool of the Schrodinger software was used to evaluate the drug-likeliness property analysis. Our selected various properties such as Molecular Weight (MW), Total solvent accessible surface; Hydrogen Bond Acceptor (HBA), Hydrogen Bond Donor (HBD), Predicted aqueous solubility (QP log S) and Human oral absorption were assessed. The lower molecular weight is an indication of better absorption of the drug compounds. A Log S value represents solubility, the lesser the Log S value means higher the solubility, which would enhance the absorption property. Lower the total solvent accessible surface was favorable for drug like properties. The chemical name of these eleven lead molecules with their corresponding Drug Bank Id and MDPI database Id and MayBridge HitFinder database Id has been given to their chemical properties (Table 3).
| S. no. | Compound Ida | Mol. Wt.b | SASAc | Human oral absorptiond | HB donore | HB acceptorf | QP log Sg |
|---|---|---|---|---|---|---|---|
| 1 | DB07075 | 384.483 | 716.679 | 54.797 | 4 | 6 | -4.593 |
| 2 | DB01940 | 474.512 | 798.629 | 67.044 | 3 | 9 | -5.017 |
| 3 | DB07025 | 386.496 | 719.51 | 71.653 | 3 | 5 | -4.284 |
| 4 | DB02224 | 304.256 | 514.08 | 52.196 | 4 | 7 | -2.67 |
| 5 | DB01094 | 302.283 | 541.774 | 75.467 | 2 | 5 | -3.782 |
| 6 | DB08009 | 414.934 | 769.425 | 94.926 | 2 | 6 | -5.745 |
| 7 | MDPI15603 | 311.34 | 566.422 | 94.53 | 2 | 5 | -3.938 |
| 8 | MDPI15661 | 311.34 | 575.036 | 92.168 | 2 | 5 | -4.084 |
| 9 | MDPI15605 | 311.34 | 563.412 | 100 | 2 | 5 | -3.887 |
| 10 | MH13092 | 290.317 | 556.494 | 96.106 | 1 | 4 | -3.922 |
| 11 | MH10963 | 302.281 | 536.82 | 75.956 | 2 | 5 | -3.699 |
aCompound Id; bMolecular weight (acceptable range is: ≤ 500)’; cTotal solvent accessible surface area (acceptable range is: 300-1000); dHuman oral absorption (acceptable range is:<25% less and >80% high); eHydrogen bond donor (acceptable range is: ≤ 5); fHydrogen bond acceptor (acceptable range is: ≤ 10); gPredicted aqueous solubility (acceptable range is: -6.5-0.5).
Table 3. Drug-like properties of the selected inhibitors from DrugBank, MDPI and MayBridge HItFinder databases.
ADMET prediction
The admetSAR server was used to find out the ADMET properties of the selected drug compounds. HIA probability, Caco-2 probability, AMES test and carcinogenicity and Rat acute toxicity prediction. HIA represent the human intestinal absorption score. The compound was having high score would be absorbed better in the intestinal tract upon oral administration. BBB represents the blood brain barrier which should be high, the higher the BBB better the penetration. AMES test was performed to find out that the compounds were mutagenic or not. We found that all the compounds showed non-toxic properties except one. Carcinogenicity also reveals that all the compounds are non-carcinogens. A compound with a lower LD50 is more lethal than the compound having higher LD50. Predicting the efflux by P-glycoprotein (P-gp) metabolism of the drug compounds is carried out by a family of microsomal enzymes known as cytochrome P450 (CYP450). The two most important members are CYP3A4 and CYP2D6 which are represented in Tables 4 and 5.
| S. no. | Ligand | BBB probability | HIA probability | Caco-2 probability | AMES test | Carcinogenicity | Rat acute toxicity (LD50: mol/kg) |
|---|---|---|---|---|---|---|---|
| 1 | DB07075 | 0.8541 | 0.9671 | 0.6352 | Non-toxic | Non-carcinogen | 2.5786 |
| 2 | DB01940 | 0.7306 | 0.9139 | 0.6892 | Non-toxic | Non-carcinogen | 2.4425 |
| 3 | DB07025 | 0.894 | 0.9647 | 0.6532 | Non-toxic | Non-carcinogen | 2.5263 |
| 4 | DB02224 | 0.5711 | 0.965 | 0.8957 | Non-toxic | Non-carcinogen | 3.02 |
| 5 | DB01094 | 0.6591 | 0.9511 | 0.8286 | Non-toxic | Non-carcinogen | 3.1455 |
| 6 | DB08009 | 0.6782 | 0.8536 | 0.59 | Non-toxic | Non-carcinogen | 2.684 |
| 7 | MDPI15603 | 0.7797 | 0.8359 | 0.7444 | Non-toxic | Non-carcinogen | 2.5097 |
| 8 | MDPI15661 | 0.8307 | 0.8628 | 0.7785 | Toxic | Non-carcinogen | 2.6928 |
| 9 | MDPI15605 | 0.7453 | 0.9394 | 0.7199 | Non-toxic | Non-carcinogen | 2.2957 |
| 10 | MH13092 | 0.8661 | 0.9743 | 0.5332 | Non-toxic | Non-carcinogen | 2.4772 |
| 11 | MH10963 | 0.6591 | 0.9511 | 0.8286 | Non-toxic | Non-carcinogen | 3.1455 |
BBB: Blood Brain Barrier; value closer to 1 represents better permeability through BBB; (2) HIA: Human Intestinal Permeability; value closer to 1 represents better absorption through intestine; (3) Caco-2: Human Intestinal Cell Line used for in-silico simulation; (4) AMES Test is done to detect a probable mutagen. A positive toxicity indicates the molecule for being a probable mutagen. (5) Carcinogenicity indicates the cancer causing ability of a molecule. (6) LD50: Lethal Dose 50 i.e., the amount of a drug which could kill 50% of the population of the organism on which it is being tested (here rat).
Table 4. In-silico absorption and toxicity profile obtained from admetSAR server.
| S. no. | Ligand | P-gp substrate/inhibitor probability | CYP-2C9 substrate/inhibitor | CYP-2D6 substrate/inhibitor | CYP-3A4 substrate/inhibitor | CYP-1A2 substrate/Inhibitor | CYP-2C19 inhibitor | CYP inhibitory promiscuity |
|---|---|---|---|---|---|---|---|---|
| 1 | DB07075 | Substrate/Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-Inhibitor | Non-Inhibitor | High |
| 2 | DB01940 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-Inhibitor | Non-Inhibitor | Low |
| 3 | DB07025 | Substrate/Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Inhibitor | substrate/Inhibitor | Inhibitor | Non-Inhibitor | High |
| 4 | DB02224 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Inhibitor | Inhibitor | Non-Inhibitor | High |
| 5 | DB01094 | Substrate/Non-Inhibitor | Non-substrate/Inhibitor | Non-substrate/Inhibitor | Non-substrate/Inhibitor | Inhibitor | Inhibitor | High |
| 6 | DB08009 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | substrate/Non-Inhibitor | Non-Inhibitor | Non-Inhibitor | Low |
| 7 | MDPI15603 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Inhibitor | substrate/Inhibitor | Inhibitor | Inhibitor | High |
| 8 | MDPI15661 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Inhibitor | substrate/Inhibitor | Inhibitor | Inhibitor | High |
| 9 | MDPI15605 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Inhibitor | Non-substrate/Inhibitor | Inhibitor | Inhibitor | High |
| 10 | MH13092 | Substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | Non-substrate/Non-Inhibitor | substrate/Non-Inhibitor | Non-Inhibitor | Non-Inhibitor | Low |
| 11 | MH10963 | Substrate/Non-Inhibitor | Non-substrate/Inhibitor | Non-substrate/Inhibitor | Non-substrate/Inhibitor | Inhibitor | Inhibitor | High |
Table 5. In-silico distribution profile obtained from admetSAR server.
Drug-target binding energy analysis against screened compounds
Prime MM-GBSA (GB stands for Generalized Born) module of the Schrodinger software is very useful for finding out the protein-ligand binding affinity. The selected protein-ligand complexes were subjected to that module. It combines OPLS Molecular Mechanics Energies (EMM), surface generalized born solvation model for polar salvation (GSGB), and a nonpolar solvation term (GNP). The total free energy of binding calculation as:
ΔG bind=G complex-(G protein+G ligand) → (1)
ΔG bind: total binding free energy of complex
G complex: total energy of the complex
G protein: energy of the receptor without ligand
G ligand: energy of the unbound ligand
Where G=EMM+GSGB+GNP → (2)
The binding free energy estimated for Drug Bank, MDPI, MayBridge HitFinder selected compounds as complexes (Table 6).
| S. no. | Compound Ida | ΔG bindb | Gevdwc | Gcould | Gcovalente | GsolGBf | GsolLipog |
|---|---|---|---|---|---|---|---|
| 1 | DB07075 | -97.78 | -52.35 | -78.22 | 8.56 | 83.69 | -55.98 |
| 2 | DB01940 | -96.74 | -55.13 | -55.51 | 10.4 | 60.24 | -53.89 |
| 3 | DB07025 | -91.61 | -42.68 | -77.7 | 8.61 | 82.52 | -59.33 |
| 4 | DB02224 | -74.52 | -36.8 | -18 | 1.23 | 18.87 | -37.46 |
| 5 | DB01094 | -93.6 | -40.84 | -31.58 | 5.15 | 18.15 | -41.49 |
| 6 | DB08009 | -102.46 | -49.06 | -39.54 | 7.65 | 48.33 | -67.83 |
| 7 | MDPI15603 | -96.2 | -45.22 | -17.23 | 3.56 | 14.07 | -48.48 |
| 8 | MDPI15661 | -104.57 | -46.48 | -30 | 4.37 | 19.7 | -48.98 |
| 9 | MDPI15605 | -78.77 | -42.3 | -11.26 | 7.27 | 17.66 | -47.99 |
| 10 | MH13092 | -79.61 | -45.64 | -10.61 | 11.38 | 11.24 | -44.34 |
| 11 | MH10963 | -80.23 | -34.82 | -25.33 | 8.19 | 16.08 | -41.19 |
aCompound Id; bFree binding energy; cVan der Waal energy; dCoulomb energy; eCovalent; energy (internal energy); fGeneralized born electro-static solvation energy; glipophilic energy (nonpolar contribution estimated by solvent accessible surface area).
Table 6. Prime MM-GBSA energy calculation result of the selected inhibitors from DrugBank, MDPI and MayBridge HItFinder databases.
Superimpose structure
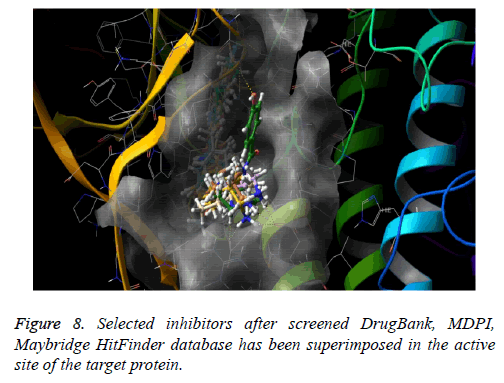
The docked pose of Drug Bank hit, MDPI hits and MayBridge HitFinder hits occupy the active binding site. It was superimposed on crystal structure (PDB ID: 4Y83) that occupy this region, The binding pattern of Drug Bank hits, MDPI hits and MayBridge HitFinder hits were found to be similar to that of the reference binding site. The figure clearly indicates that the proposed binding pose is well occupied in the active site of COT kinase complex and has a binding pattern similar to that of reference ligand (Figure 8).
Conclusion
Identification of the novel and potential lead compound is step by step procedure. A range of computational tools and techniques were used to discover best inhibitors. A very large number of drugs library were subjected for the screening against the target compound (COT kinase Complex). This structure based virtual screening helped us to reduce the number of compounds which shows the greatest binding affinity to the target and showing the best inhibitory properties. Drug likeliness properties calculated by the Lipinski filter, after using virtual screening and docking using Schrodinger software, the best 11 compounds found promising. Among all compound 3-(5-{[4-(aminomethyl) piperidin-1-yl] methyl}-1hindol- 2-yl)-1h-indazole-6-carbonitrile was found to be more potent and selective. These all compounds showing the binding where the reference co-crystallized ligand bind in the hinge region. It is obvious that these hits could be as potent and selective anticancer agent of Cancer Osaka Thyroid Kinase complex.
Acknowledgements
The Research work was carried out in the Department of Bioinformatics, Maulana Azad National Institute of Technology, India. Authors wish to acknowledge the Schrodinger team for providing software facility.
Funding
Author doesn't have any funding from the institute.
References
- Roskoski R. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res 2015; 100: 1-23.
- London CA. Kinase dysfunction and kinase inhibitors. Vet Dermatol 2013; 24: 181-187.
- Lee HW, Choi HY, Joo KM, Nam DH. Tumor progression locus 2 (Tpl2) kinase as a novel therapeutic target for cancer: double-sided effects of Tpl2 on cancer. Int J Mol Sci 2015; 16: 4471-4491.
- Beinke S, Robinson MJ, Hugunin M, Ley SC. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IkappaB kinase-induced proteolysis of NF-kappaB1 p105. Mol Cell Biol 2004; 24: 9658-9567.
- Gantke T, Sriskantharajah S, Sadowski M, Ley SC. IΚB kinase regulation of the TPL-2/ERK MAPK pathway. Immunol Rev 2012; 246: 168-182.
- Gantke T, Sriskantharajah S, Ley SC. Regulation and function of TPL-2, an IΚB kinase-regulated MAP kinase kinase kinase. Cell Res 2011; 21: 131-145.
- Lee J, Jeong S, Park JH, Lee CR, Ku CR, Kang SW. Aberrant expression of COT is related to recurrence of papillary thyroid cancer. Medicine (Baltimore) 2015; 94: e548.
- Kim JY, Lim SC, Kim G, Yun HJ, Ahn SG, Choi HS. Interleukin-33/ST2 axis promotes epithelial cell transformation and breast tumorigenesis via upregulation of COT activity. Oncogene 2015; 34: 4928-4938.
- Kim G, Khanal P, Kim JY, Yun HJ, Lim SC. COT phosphorylates prolyl-isomerase Pin1 to promote tumorigenesis in breast cancer. Mol Carcinog 2015; 54: 440-448.
- Krcova Z, Ehrmann J, Krejci V, Eliopoulos A, Kolar Z. Tpl-2/Cot and COX-2 in breast cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2008; 152: 21-25.
- Sourvinos G, Tsatsanis C, Spandidos DA. Overexpression of the Tpl-2/Cot oncogene in human breast cancer. Oncogene 1999; 18: 4968-4973.
- Gruosso T, Garnier C, Abelanet S, Kieffer Y, Lemesre V, Bellanger D. MAP3K8/TPL-2/COT is a potential predictive marker for MEK inhibitor treatment in high-grade serous ovarian carcinomas. Nat Commun 2015; 6: 8583.
- Sriskantharajah S, Guckel E, Tsakiri N, Kierdorf K, Brender C. Regulation of experimental autoimmune encephalomyelitis by TPL-2 kinase. J Immunol 2014; 192: 3518-3529.
- Ballak DB, van Essen P, van Diepen JA, Jansen H, Hijmans A, Matsuguchi T. MAP3K8 (TPL2/COT) affects obesity-induced adipose tissue inflammation without systemic effects in humans and in mice. PLoS One 2014; 9: e89615.
- Sanz-Garcia C, Ferrer-Mayorga G, Gonzalez-Rodriguez A, Valverde AM, Martin-Duce A, Velasco-Martin JP. Sterile inflammation in acetaminophen-induced liver injury is mediated by Cot/tpl2. J Biol Chem 2013; 288: 15342-15351.
- Lee HW, Joo KM, Lim JE, Cho HJ, Cho HJ. Tpl2 kinase impacts tumor growth and metastasis of clear cell renal cell carcinoma. Mol Cancer Res 2013; 11: 1375-1386.
- Jeong JH, Bhatia A, Toth Z, Oh S, Inn KS, Liao CP. TPL2/COT/MAP3K8 (TPL2) activation promotes androgen depletion-independent (ADI) prostate cancer growth. PLoS One 2011; 6: 16205.
- Jager J, Gremeaux T, Gonzalez T, Bonnafous S, Debard C, Laville M. Tpl2 kinase is upregulated in adipose tissue in obesity and may mediate interleukin-1beta and tumor necrosis factor-{alpha} effects on extracellular signal-regulated kinase activation and lipolysis. Diabetes 2010; 59: 61-70.
- Van Acker GJ, Perides G, Weiss ER, Das S, Tsichlis PN. Tumor progression locus-2 is a critical regulator of pancreatic and lung inflammation during acute pancreatitis. J Biol Chem 2007; 282: 22140-22149.
- Hu Y, Cole D, Denny RA, Anderson DR, Ipek M. Discovery of indazoles as inhibitors of Tpl2 kinase. Bioorg Med Chem Lett 2011; 21: 4758-4761.
- Cusack K, Allen H, Bischoff A, Clabbers A, Dixon R. Identification of a selective thieno[2,3-c]pyridine inhibitor of COT kinase and TNF-alpha production. Bioorg Med Chem Lett 2009; 19: 1722-1725.
- George D, Friedman M, Allen H, Argiriadi M, Barberis C. Discovery of thieno[2,3-c]pyridines as potent COT inhibitors. Bioorg Med Chem Lett 2008; 18: 4952-4955.
- Kaila N, Green N, Li HQ, Hu Y, Janz K, Gavrin LK. Identification of a novel class of selective Tpl2 kinase inhibitors: 4-Alkylamino-[1,7]naphthyridine-3-carbonitriles. Bioorg Med Chem 2007; 15: 6425-6442.
- Green N, Hu Y, Janz K, Li HQ, Kaila N, Guler S. Inhibitors of tumor progression loci-2 (Tpl2) kinase and tumor necrosis factor alpha (TNF-alpha) production: selectivity and in vivo antiinflammatory activity of novel 8-substituted-4-anilino-6-aminoquinoline-3-carbonitriles. J Med Chem 2007; 50: 4728-4745.
- Gavrin LK, Green N, Hu Y, Janz K, Kaila N, Li HQ. Inhibition of Tpl2 kinase and TNF-alpha production with 1,7-naphthyridine-3-carbonitriles: synthesis and structure-activity relationships. Bioorg Med Chem Lett 2005; 15: 5288-5292.
- Arrington K, Fraley M, Garbaccio R, Huang S, Lindsley C, Steen J. Inhibitors of Checkpoint Kinases. Google Patents 2008.
- Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol Oncol 2012; 6: 155-176.
- Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science 2004; 303: 1800-1805.
- McInnes C. Virtual screening strategies in drug discovery. Curr Opin Chem Biol 2007; 11: 494-502.
- Gutmann S, Hinniger A, Fendrich G, Druckes P, Antz S, Mattes H. The crystal structure of cancer osaka thyroid kinase reveals an unexpected kinase domain fold. J Biol Chem 2015; 290: 15210-15218.
- Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J Comput Chem 2005; 26: 1752-1780.
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 2006; 49: 6177-6196.
- Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 1993; 98: 5648-5652.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001; 46: 3-26.
- Yamashita F, Hashida M. In silico approaches for predicting ADME properties of drugs. Drug Metab Pharmacokinet 2004; 19: 327-338.
- Lyne PD, Lamb ML, Saeh JC. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem 2006; 49: 4805-4808.