ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 1
Molecular mechanism of miR-378 in enhancing the invasion and migration of hepatoma carcinoma cells through targeted PTEN-based EMT regulation
Yuexiang Niu*
Department of Infectious Diseases, Linyi Central Hospital, Linyi, Shandong, PR China
- *Corresponding Author:
- Yuexiang Niu
Department of Infectious Diseases
Linyi Central Hospital, PR China
Accepted date: October 26, 2017
DOI: 10.4066/biomedicalresearch.29-17-2885
Visit for more related articles at Biomedical ResearchObjective: We aimed to discuss the molecular mechanism of miR-378 in enhancing the invasion and migration of hepatoma carcinoma cells through targeted PTEN-based EMT regulation.
Method: The human hepatoma cell line HepG2 was cultured. Cells in the logarithmic phase were collected and transfected using miR-378 mimics through liposome-mediated transfection method. Successfully transfected cells and the primary cancer cells were denoted as groups A and B. Each group had eight repeated holes. The invasion and migration abilities of both groups were tested by Transwell and wound-healing experiments at 24 and 48 h. The relative miR-378 mRNA expression levels of both groups were detected by reverse transcription-polymerase chain reaction. PTEN and EMT related proteins were tested by Western blot at 48 h. Differences between two groups were analysed by SPSS20.0.
Results: The cell-leaching and wound-healing rates of group A at 24 and 48 h were lower than those of group B. Cell-leaching and wound-healing rates at 48 h were higher than those at 24 h. Differences were statistically significant (P<0.05). The relative expression levels of miR-378 mRNA and PTEN protein content of group A were significantly higher than those of group B, whereas relative expression levels of Protein Kinase B (PKB), Ki67 and Phosphatidylinositol 3 (PI3K) were significantly lower (P<0.05).
Conclusions: Low expression of miR-378 was observed in hepatoma carcinoma cells, which can downregulates the expression of PTEN protein and upregulates the expression of EMT-related proteins, thereby enhancing their invasion and migration abilities.
Keywords
miR-378, PTEN, EMT, Liver cancer, Invasion, Migration
Introduction
Liver cancer is a common malignant tumor that inflicts serious damage. In China, the morbidity rates of this disease are high. Hepatoma carcinoma cells reportedly have strong invasion and migration abilities. Liver-cancer patients easily suffer relapse, as well as hepatoma carcinoma cell migration and infiltration, which are life-threatening [1]. Thus, discussing the regulatory mechanism of hepatoma carcinoma cell invasion and migration is crucial. miRNAs are noncoding micromolecule RNAs that inhibit gene expression after transcription and are closely related to uncontrollable cell proliferation, invasion, and transfer regulation. Research has reported that the expression level of miR-378 in liver-cancer tissues is significantly lower than that in normal tissues near the cancer [2]. It is related with clinical stages, lymphatic metastasis, prognosis, and long-term survival rate. However, only few studies concerning the regulatory mechanism of miR-378 in hepatoma carcinoma cell invasion and migration have been reported. Meanwhile, PTEN can regulate EMT-related proteins closely related to the invasion and migration of malignant tumors [3]. Evidence has shown that miR-378 may regulate PTEN in cancer cells. In the present study, we envisaged that miR-378 may regulate PTEN and EMT-related proteins to enhance the invasion and migration abilities of hepatoma carcinoma cells. An in vitro experiment was conducted for verification.
Materials and Methods
Materials
The human hepatoma cell line HepG2 was purchased from Nanjing Jiancheng Biological Engineering Institute. Fetal calf serum, culture medium, and trypsin were all from Shanghai Fuxiang Biotechnology Co., Ltd. miR-378 mimics sequence, miR-378 primer sequence, and internal reference sequence were designed and synthesized by Shenzhen Jingmei Biotechnology Co., Ltd. CO2 incubator and centrifugal machine were from Hunan Xiangyi Instrument Factory. Electrophoresis apparatus and gel-imaging analysis system were from Bio-Rad Company (USA). Micropipettor was from Eppendorf Company (Germany). The clean bench was from Suzhou Antai Air Technique Co., Ltd. Fluorogenic quantitative PCR instrument was from Beijing 61 Instrument Plant.
Methods
Cell-invasion ability was tested by Transwell experiment. Transwell cells were placed on a piece of culture plate, forming one top and one bottom chamber. A total of 50 μL of culture solution diluted by serum-free medium was paved uniformly on the upper chamber, and the culture plate was placed in the incubator (37°C). The cell concentrations of both groups were adjusted to 2.5 × 105/ml by serum-free medium. Then 100 μL of cell suspension was dripped to the top chamber, and 500 μL of culture solution was added to the bottom chamber. The plate was placed in the incubator and cultured under 37°C and 5% CO2. Cells in the top chamber were eliminated with a cotton swab at 24 and 48 h and then fixed with 4% formaldehyde solution. Conventional dying was observed under an optical microscope. Cell-leaching rate was calculated by randomly choosing five views of 200 amplifications. The test was repeated three times, and the mean value was used.
Cell-migration ability was tested by wound-healing experiment [4]. Two groups of cells were inoculated onto a plate with 96 holes at 2 × 106 cells per hole. A hungry culture was implemented for 12 h when cells reached 90% convergence. Wounds were then made by pipette, and cells were sequentially rinsed for three times with PBS buffer solution. Cells in wounds were added with serum-free culture medium and cultured in the incubator according to the above conditions. Wound distances at 24 and 48 h were measured and photographed. Wound-healing rate, i.e., the ratio between wound-width changes and wound width before the culture, was calculated three times, and mean results were used.
The expression levels of PTEN and EMT-related proteins were determined by Western blot analysis [5]. The total proteins of both groups were extracted with a kit. After protein concentrations were determined, the relative expression levels of the above proteins were tested. First, SDS-PAGE gel was prepared. Second, test samples were evenly mixed with buffer solution. Conventional transmembrane was performed after the gel test. The corresponding first and second antibodies were added successively after being enclosed for incubation. Samples were developed and photographed later, and then stripes were compared. The relative expression levels of proteins were tested using software.
Observation indices
The test results of both groups in invasion and migration abilities at 24 and 48 h were compared to obtain cell-leaching and wound-healing rates. The relative expression levels of miR-378 mRNA (2-ΔΔCT) of both groups were compared. The relative expression levels of PTEN and EMT-related proteins of both groups were compared to obtain the optical density of PTEN, PKB, Ki67, and PI3K.
Statistical analysis
Statistical analysis was accomplished by SPSS20.0. Measurement data (͞x ± s) and enumeration data (%) were tested using the t and χ2 tests. P<0.05 means statistically significant difference.
Results
Comparison of invasion and migration abilities
The cell-leaching and wound-healing rates of group A at 24 and 48 h were lower than those of group B. For both groups, cell-leaching and wound-healing rates at 48 h were higher than those at 24 h. The inter- and intra-group differences were statistically significant (P<0.05) (Table 1).
| Groups | n | Leaching rate (%) | t | P | Wound-healing rate (%) | t | P | ||
|---|---|---|---|---|---|---|---|---|---|
| After 24 h | After 48 h | After 24 h | After 48 h | ||||||
| Group A | 8 | 18.8 ± 3.3 | 32.0 ± 5.1 | 6.146 | 0.000 | 15.4 ± 2.1 | 28.4 ± 3.5 | 9.008 | 0.000 |
| Group B | 8 | 29.5 ± 3.1 | 54.6 ± 5.4 | 11.402 | 0.000 | 20.1 ± 2.9 | 43.6 ± 3.7 | 14.139 | 0.000 |
| t | 6.684 | 8.606 | / | / | 3.713 | 8.441 | / | / | |
| P | 0.000 | 0.000 | / | / | 0.002 | 0.000 | / | / | |
Table 1. Comparison on cell-leaching and wound-healing rates of groups A and B (͞x ± s).
Comparison of the relative expression of miR-378 mRNA
The relative expression of miR-378 mRNA of group A was significantly higher than that of group B (P<0.05) (Table 2).
| Groups | n | Relative expression of miR-378 mRNA |
|---|---|---|
| Group A | 8 | 0.378 ± 0.019 |
| Group B | 8 | 0.181 ± 0.010 |
| t | 25.951 | |
| P | 0.000 |
Table 2. Comparison on the relative expression of miR-378 mRNA between groups A and B (͞x ± s).
Comparison of the relative expression of PTEN and EMT-related proteins
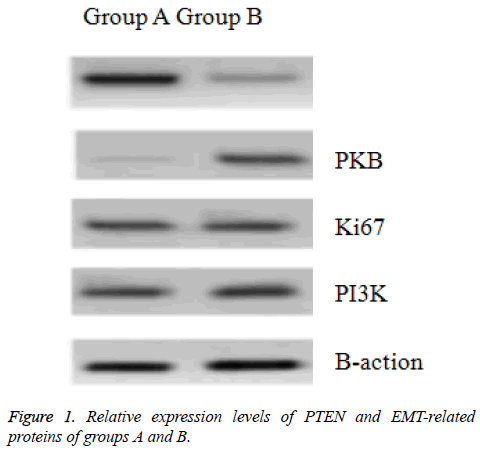
The PTEN content of Group A was far higher than that of group B, but relative expression levels of PKB, Ki67, and PI3K were significantly lower (P<0.05) (Table 3 and Figure 1).
| Groups | n | PTEN | PKB | Ki67 | PI3K |
|---|---|---|---|---|---|
| Group A | 8 | 0.983 ± 0.014 | 0.217 ± 0.026 | 0.520 ± 0.020 | 0.697 ± 0.019 |
| Group B | 8 | 0.456 ± 0.016 | 0.769 ± 0.021 | 0.781 ± 0.025 | 0.805 ± 0.018 |
| t | 70.111 | 46.715 | 23.058 | 11.671 | |
| P | 0.000 | 0.000 | 0.000 | 0.000 |
Table 3. Comparison of relative expression levels of PTEN and EMTrelated proteins between groups A and B (͞x ± s).
Discussion
In this work, we discovered that the cell-leaching and woundhealing rates of group A were significantly lower than those of group B at any time. Group A were cells transfected by miR-378 mimics, and group B were original hepatoma carcinoma cells. The RT-PCR test results of group A indicated that the relative expression level of the miR-378 mRNA of group A were far higher than that of group B, showing that miR-378 expression in hepatoma carcinoma cells was inhibited. This finding may be related to the enhanced invasion and migration abilities of miR-378. The author has reviewed recent associated studies but discovered no identical research methods and conclusions. However, a foreign study has shown that miR-378 can indeed enhance the invasion and migration abilities of HepG2. Another work has demonstrated that the relative expression of miR-378 mRNA in the blood cells of patients with viral hepatitis due to liver cancer is far lower than that in normal healthy people, implying that abnormal miR-378 expression may be closely correlated with liver-cancer incidence [6,7]. These findings require further exploration.
Research on PTEN regulation over EMT-related proteins started early. At the end of the 20th century, PTEN was cloned and became the earliest discovered cancer-suppressor gene with phosphatase activity. As a part of the PI3K/AKT signal path, PTEN plays an important role in a series of biological changes, such as cell survival, growth, proliferation, invasion, migration, and differentiation [8]. Under the regulatory effect of PETN, abnormal expression levels of PKB, Ki67 and PI3K intensify, resulting in cell depolarization and sharply weakening adhesive force. Consequently, cell invasion and migration abilities are enhanced, as manifested by cachexia. This regulatory mechanism has been verified in cells of various diseases, such as gliomas, Hodgkin lymphoma, and hematological malignant tumor [9]. Researchers reported that a low expression of PTEN in hepatoma carcinoma cells, which weakens the inhibition of EMT-related proteins, thereby significantly enhancing cell proliferation, invasion, and migration [10].
The present study further revealed that PTEN expression in group A was higher than that in group B, but the relative expression levels of PKB, Ki67 and PI3K were lower. Thus, PTEN had low expression levels in hepatoma carcinoma cells, whereas PKB, Ki67and PI3K had high relative expression levels. Combining the cell-invasion and migration test results, as well as the relative expression-test results of miR-378 mRNA, of both groups, we speculated that low miR-378 expression can reduce PTEN content and enhance the activity of EMT-related proteins, thereby facilitating hepatoma carcinoma cell invasion and migration.
Conclusion
In sum, miR-378 had low expression levels in hepatoma carcinoma cells, which can regulate the expression levels of PTEN and EMT-related proteins and thus enhance hepatoma carcinoma cell invasion and migration. Research conclusions provide a new idea and action targets. However, whether the miR-378 regulation of expression levels of other genes and proteins influenced hepatoma carcinoma cell invasion and migration, as well as the specific underlying mechanism, requires further research.
References
- Li S, Yang F, Wang M. miR-378 functions as an onco-miRNA by targeting the ST7L/Wnt/β-catenin pathway in cervical cancer. Int J Mol Med 2017; 40: 1047-1056.
- Pizzamiglio S, Zanutto S, Ciniselli CM. A methodological procedure for evaluating the impact of hemolysis on circulating microRNAs. Oncol Lett 2017; 13: 315-320.
- Kuang X, Wei C, Zhang T. miR-378 inhibits cell growth and enhances apoptosis in human myelodysplastic syndromes. Int J Oncol 2016; 49: 1921-1930.
- Mirzaei H, Khataminfar S, Mohammadparast S. Circulating microRNAs as potential diagnostic biomarkers and therapeutic targets in gastric cancer: current status and future perspectives. Curr Med Chem 2016; 23: 4135-4150.
- Dhayat SA, Mardin WA, Seggewiß J. MicroRNA profiling implies new markers of gemcitabine chemoresistance in mutant p53 pancreatic ductal adenocarcinoma. PLoS One 2015; 10: 107-112.
- Weng WH, Leung WH, Pang YJ. Lauric acid can improve the sensitization of Cetuximab in KRAS/BRAF mutated colorectal cancer cells by retrievable microRNA-378 expression. Oncol Rep 2016; 35: 107-116.
- Ma J, Lin J, Qian J. MiR-378 promotes the migration of liver cancer cells by down-regulating Fus expression. Cell Physiol Biochem 2014; 34: 2266-2274.
- Zhu XW, Wen XM, Zhang YY. The 5 flanking region of miR-378 is hypomethylated in acute myeloid leukemia. Int J Clin Exp Pathol 2015; 8: 4321-4331.
- Li B, Wang Y, Li S. Decreased expression of miR-378 correlates with tumor invasiveness and poor prognosis of patients with glioma. Int J Clin Exp Pathol 2015; 8: 7016-7021.
- Chen QG, Zhou W, Han T. MiR-378 suppresses prostate cancer cell growth through downregulation of MAPK1 in vitro and in vivo. Tumour Biol 2016; 37: 2095-2103.