ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 10
Molecular probes for identification of UDP-glucuronosyltransferase 1 gene polymorphisms for Gilbert's syndrome diagnosis
Xuping Wu1*, Dongrui Zhou2, Jinyun Song1, Mei Sun1 and Yan Pan3*
1The Second Hospital of Nanjing Affiliated to Southeast University, Nanjing, PR China
2Key Laboratory of Child Development and Learning Science, Southeast University, Nanjing, PR China
3Lianshui County People’s Hospital, Jiangsu, PR China
- *Corresponding Authors:
- Yan Pan
Lianshui County
People’s Hospital Jiangsu, PR China
Xuping Wu
The Second Hospital of Nanjing
Affiliated to the Medical School of Southeast University
PR China
Accepted date: April 03, 2018
DOI: 10.4066/biomedicalresearch.29-18-456
Visit for more related articles at Biomedical ResearchThe T-3279G mutation in Phenobarbital Responsive Enhancer Modulein (PBREM), TA-insertion in the TATA box, creating the A(TA)7TAA motif instead of the A(TA)6TAA, and G211A mutation in coding exon 1, particularly in Asians, of human UGT1A1 (UDP-glucoronosyltransferase) gene are the three common polymorphisms of Gilbert’s syndrome. This article employed dot blot hybridization to detect the T-3279G, A(TA)6/7TAA and G211A mutation of UGT1A1 gene in 15 patients. In the dot blot hybridization assay, PCR products were denatured and spotted on positively charged nylon membranes instead of acrylamide-modified slides. Then three pairs of probes were labeled by digoxigenin and hybridized with the spotted array. After hybridization, the signal was visualized using TMB color development solution and all possible polymorphisms of the T-3279G, A(TA)6/7TAA and G211A in 15 Chinese patients with hyperbilirubinemia were successfully identified. The results of 15 samples by dotblot hybridization were consistent with the sequencing results. The data showed that dot blot hybridization assay is a quick and low cost method to detect UGT1A1 gene polymorphisms and helpful to diagnosis of Gilbert’s syndrome.
Keywords
UDP-glucuronosyltransferase 1A1, Molecular probes, Dot blot hybridization, Polymorphisms
Introduction
UDP-glucuronosyltransferases (UGTs), one important family of phase II drug metabolizing enzymes, catalyze the biotransformation of many endobiotics and xenobiotics, and are coded by polymorphic genes [1]. UGT enzymes are mainly expressed in the liver and the intestine and classified into different families, subfamilies and genes. UGT1A1, a member of UGT1 subfamily, is a critical enzyme in the elimination pathway of bilirubin. It is well known that inhibition of UGT1A1-mediated bilirubin clearance may lead to the elevated level of unconjugated bilirubin in serum, which may lead to hyperbilirubinemia, jaundice, hepatotoxicity and neurotoxicity. Gilbert’s syndrome is a mild hereditary unconjugated hyperbilirubinemia due to defect of 1UGT1A1. Individuals with Gilbert's syndrome exhibit lower glucuronidation rates when medicated with several HIV drugs and irinotecan [2-6].
Numerous polymorphisms of UGT1A1 have been identified [7,8]. Firstly, a Phenobarbital Responsive Enhancer Module (PBREM) is located approximately 3.5 kbp upstream from the coding region of UGT1A1 gene and the T-3279G mutation in PBREM is a common polymorphism causing Gilbert syndrome [9]. Secondly, the UGT1A1*28 polymorphism is characterized by the TA-insertion in the TATA box, creating the A(TA)7TAA motif instead of the A(TA)6TAA. This thymine- and adenine-rich regulatory region is the major recognition point for the binding of transcription factors and RNA polymerase in many genes. Clinical studies have demonstrated a significant correlation between the UGT1A1*28 polymorphism and the pharmacokinetics of drugs [10-12]. Finally, a significant association was observed between prolonged jaundice in male infants and UGT1A1 G71R polymorphism (G211A mutation in coding exon 1 of UGT1A1 gene) [13]. The coding region polymorphism is found only in Asians and is associated with decreased enzyme function, whereas the promoter polymorphism is associated with decreased gene expression. Individuals carriers of the UGT1A1*6 (G211A mutation) and *28 variants are apparently much more likely to develop jaundice after medication with methotrexate, indinavir, sorafenib, atazanavir or other drugs inhibiting UGT1A1. This is presumably due to the lower functional reserve available to compensate UGT1A1 inhibition [6].
To our knowledge, no study has analyzed the relationship between UGT1A1 polymorphisms and dot blot hybridization assay with molecular probes. In the dot blot hybridization assay, the obtained PCR products were successively purified denatured, and then digoxigenin-labeled probes were allowed to hybridize to DNA array. The hybridization results could be determined by visualizing the DNA array using bioluminesce method.
Previously, we applied three dimensional acrylamide gel method to detect UGT1A1 gene polymorphism [14]. However, preparation of the acrylamide-modified slides is timeconsuming. Fluorescent labeled probes, acrylamide-modified primers and the use of Luxscan-10K/A (CapitalBio Company, China) are expensive. Other method, such as direct sequencing method, TaqMan MGB SNP genotyping assay, DNA melting curve analysis, Restriction Fragment Length Polymorphism (RFLP) method have been used to detect mutations of UGT1A1 gene for Gilbert’s Syndrome diagnosis [15-17]. In this study, we applied dot blot hybridization assay with molecular probes for the first time to detect the T-3279G, A(TA)6/7TAA and G211A mutation of UGT1A1 gene.
Materials and Methods
Participants and DNA isolation
15 Chinese patients with hyperbilirubinemia were recruited at the Second Affiliated Hospital of Southeast University, Nanjing, P. R. China. All patients had total bilirubin levels (TBIL) of ≥ 220.6 umol/L, and no signs or symptoms indicative of other hepatobiliary diseases. Serum samples were used for the evaluation of total bilirubin level, and all biochemical tests were carried out in the same laboratory with standardized laboratory methods. Peripheral blood samples were collected from all participants in the morning after overnight fasting. Total DNA was extracted using the QIAamp DNA midi blood kit (QIAGEN, Germany) according to the standard protocol. All participants provided a written informed consent before enrolment and all research procedures were approved by the Ethics Committee of the Second Affiliated Hospital of Southeast University.
PCR amplification
A pair of primers F1 (5’-CACCTCCTCCTTATTCTCTT-3’) and R1 (5’-CTCATTCCTCCTCTCTAGCC-3’), whose design was based on published DNA sequence, was used in the PCR to obtain the PBREM region of UGT1A1 gene. The procedure was as follows: 94°C for 3 min, 32 cycles of 94°C for 30 s, 54.2°C for 45 s, and 72°C for 45 s, and then 72°C for 10 min. The region containing TATA-box and 211 site of UGT1A1 gene was generated by PCR using Ex Taq (Takara, Japan) with two primers F2 (5’-CCCTGCTACCTTTGTGGACT-3’) and R2 (5’-CATTATGCCCGAGACTAACAAA -3’). The reaction conditions were 94°C for 3 min followed by 32 cycles of 94°C for 30 s, 57°C for 45 s, and 72°C for 45 s, and then a final elongation step at 72°C for 10 min. After agarose gel electrophoresis analysis, PCR products were processed by ethanol precipitation overnight at -20°C. Then, PCR products were harvested by centrifugation at 14000 g for 20 min and diluted in water.
PCR products immobilization
PCR products were denatured by mixing with the same volume of denaturation solution (0.5 N NaOH, 1.5 M NaCl) at room temperature for 30 min. The denatured DNAs were then spotted on 25 mm positively charged nylon membranes (Roche Diagnostics, Mannheim, Germany), followed by baking at 100°C for 30 min.
Hybridization with the corresponding probes
The spotted array was pre-hybridized with 1 × DIG Easy Hyb (Roche Diagnostics) in a hybridization oven (Robbins Scientific Corp., USA) for 4 h at 60°C. Samples of 10 ml (50 ng) of DIG-labeled candidate probe (Table 1) was denatured for 10 min at 98°C and mixed with 1 × DIG Easy Hyb (1.5 ml) warmed to 60°C. The mixture was incubated with nylon membranes bearing DNA arrays overnight at 37°C in the hybridization overnight.
| Probe | Probe sequences |
|---|---|
| -3279T | 5’-Digoxigenin-TTCAGTTTGAACA-3’ |
| -3279G | 5’-Digoxigenin-TTCAGTGTGAACA-3’ |
| A(TA)6TAA | 5’-Digoxigenin-GCCATATATATATATATAAG-3’ |
| A(TA)7TAA | 5’-Digoxigenin-GCCATATATATATATATATAAG-3’ |
| 211G | 5’-Digoxigenin-AGAG ACGGAGCAT-3’ |
| 211A | 5’-Digoxigenin-AGAGACAGAGCAT-3’ |
| Bold under led characters indicate the test loci. | |
Table 1. The probe sequences used in this study.
Image scanning
After hybridization, the nylon membranes were washed twice with 50 ml of 23 SSC/0.1% SDS at room temperature, each for 10 min, and twice with 50 ml of 0.53 SSC/0.1% SDS at 62°C, each for 10 min, respectively. Then the nylon membranes were blocked with 10% BSA for 1 h. The membranes were incubated with a 1:1000 dilution of Horseradish Peroxidaseconjugated IgG Fraction Monoclonal Mouse Anti-Digoxin Antibody (Jackson, USA) for 30 min and then washed three times with TBST buffer (20 mM Tris HCl pH 8.0, 150 mM NaCl, and 0.05% (v/v) Tween 20). Finally, the signal was visualized using TMB color development solution (Phygene, China).
Results
The general clinical data are shown in Table 2. 15 patients included 10 males and 5 females with a median age of 55 y (range, 38-68). All patients had TBIL of ≥ 220.6 umol/L. In principle, two rounds of dot-blot hybridization were carried out simultaneously.
| Patient | Gender | Age | TBIL (μmol/L) | DBIL (μmol/L) | IBIL (μmol/L) |
|---|---|---|---|---|---|
| 1 | Male | 56 | 300.4 | 36 | 264.4 |
| 2 | Male | 60 | 287.1 | 15.6 | 271.5 |
| 3 | Male | 48 | 252 | 9.5 | 242.5 |
| 4 | Female | 45 | 312.5 | 33.2 | 279.3 |
| 5 | Female | 68 | 227.1 | 6 | 221.1 |
| 6 | Male | 43 | 280 | 12.6 | 267.4 |
| 7 | Male | 62 | 336.8 | 38 | 298.8 |
| 8 | Female | 53 | 325 | 35 | 290 |
| 9 | Male | 66 | 246.9 | 5.9 | 241 |
| 10 | Male | 55 | 288.1 | 22.3 | 265.8 |
| 11 | Female | 58 | 252 | 7.9 | 244.1 |
| 12 | Female | 47 | 308.4 | 22.3 | 286.1 |
| S1 | Male | 61 | 273.3 | 13 | 260.3 |
| S2 | Male | 56 | 268.9 | 8.7 | 260.2 |
| S3 | Male | 38 | 319.6 | 30.9 | 288.7 |
Table 2. General clinical data in the 15 patients.
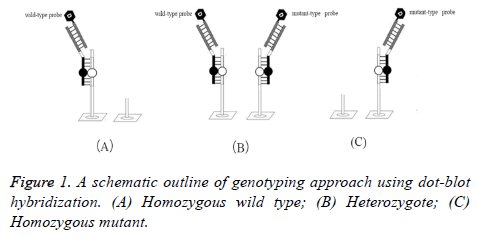
The homozygous wild type hybridized with only digoxigeninlabeled wild-type probes, showing wild-type signals. While the homozygous mutant hybridized with only digoxigenin-labeled mutant-type probes, showing mutant signals. Both the wildtype signals and the mutant signals were shown for the heterozygote. So, mutant alleles were distinguished from wildtype alleles. A schematic outline of genotyping approach using dot-blot hybridization is shown in Figure 1.
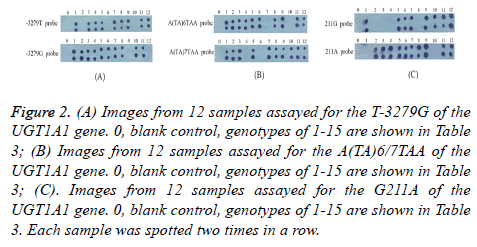
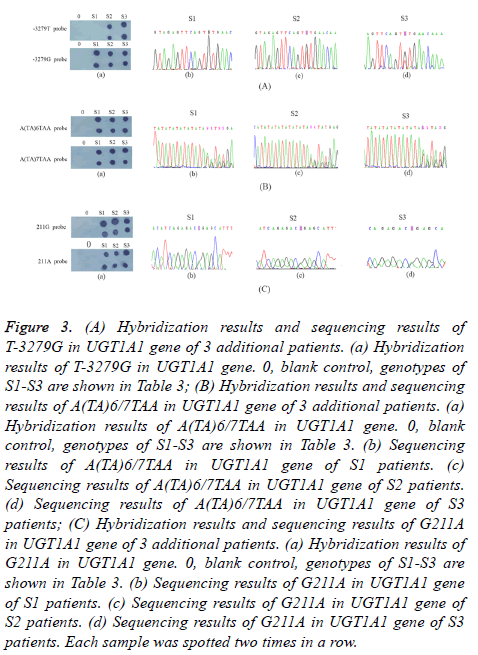
12 samples were screened by dot-blot hybridization. All possible genotypes of 12 samples were successfully identified and shown in Figure 2. Images from 12 samples assayed for the T-3279G, A(TA)6/7TAA, G211A of the UGT1A1 were shown in Figures 2 (A)-2(C), respectively. In addition, 3 patients were also screened by dot-blot hybridization and sequencing results of the PCR products were shown in Figure 3. Hybridization results and sequencing results of T-3279G, A(TA)6/7TAA, G211A in UGT1A1 gene of 3 additional patients were shown in Figures 3(A)-3(C), respectively. Variations of UGT1A1 genotypes in 15 patients are shown in Table 3.
Figure 2: (A) Images from 12 samples assayed for the T-3279G of the UGT1A1 gene. 0, blank control, genotypes of 1-15 are shown in Table 3; (B) Images from 12 samples assayed for the A(TA)6/7TAA of the UGT1A1 gene. 0, blank control, genotypes of 1-15 are shown in Table 3; (C). Images from 12 samples assayed for the G211A of the UGT1A1 gene. 0, blank control, genotypes of 1-15 are shown in Table 3. Each sample was spotted two times in a row.
Figure 3: (A) Hybridization results and sequencing results of T-3279G in UGT1A1 gene of 3 additional patients. (a) Hybridization results of T-3279G in UGT1A1 gene. 0, blank control, genotypes of S1-S3 are shown in Table 3; (B) Hybridization results and sequencing results of A(TA)6/7TAA in UGT1A1 gene of 3 additional patients. (a) Hybridization results of A(TA)6/7TAA in UGT1A1 gene. 0, blank control, genotypes of S1-S3 are shown in Table 3. (b) Sequencing results of A(TA)6/7TAA in UGT1A1 gene of S1 patients. (c) Sequencing results of A(TA)6/7TAA in UGT1A1 gene of S2 patients. (d) Sequencing results of A(TA)6/7TAA in UGT1A1 gene of S3 patients; (C) Hybridization results and sequencing results of G211A in UGT1A1 gene of 3 additional patients. (a) Hybridization results of G211A in UGT1A1 gene. 0, blank control, genotypes of S1-S3 are shown in Table 3. (b) Sequencing results of G211A in UGT1A1 gene of S1 patients. (c) Sequencing results of G211A in UGT1A1 gene of S2 patients. (d) Sequencing results of G211A in UGT1A1 gene of S3 patients. Each sample was spotted two times in a row.
| Patient | T-3279G | A(TA)6/7TA | G211A |
|---|---|---|---|
| 1 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | homozygote G211G |
| 2 | homozygote G-3279G | heterozygote A(TA)6/7TAA | homozygote A211A |
| 3 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | homozygote A211A |
| 4 | heterozygote T-3279G | homozygote A(TA)6TAA | homozygote A211A |
| 5 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | heterozygote G211A |
| 6 | homozygote G-3279G | homozygote A(TA)6TAA | heterozygote G211A |
| 7 | heterozygote T-3279G | homozygote A(TA)7TAA | heterozygote G211A |
| 8 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | homozygote A211A |
| 9 | homozygote T-3279T | heterozygote A(TA)6/7TAA | heterozygote G211A |
| 10 | homozygote G-3279G | homozygote A(TA)7TAA | homozygote G211G |
| 11 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | heterozygote G211A |
| 12 | homozygote T-3279T | heterozygote A(TA)6/7TAA | heterozygote G211A |
| S1 | homozygote G-3279G | heterozygote A(TA)6/7TAA | heterozygote G211A |
| S2 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | heterozygote G211A |
| S3 | heterozygote T-3279G | heterozygote A(TA)6/7TAA | heterozygote G211A |
Table 3. Variations of UGT1A1 genotypes in 15 patients.
Discussion
UGT1A1 is the only physiologically relevant UGT isoform involved in the metabolic clearance of endobiotics bilirubin. Potent inhibition of UGT1A1 may result in partial or complete loss of UGT1A1 activity, and then increase the exposure of bilirubin and other endogenous (e.g., steroid hormones and bile acids) and xenobiotics (e.g., drugs, chemical carcinogens, and environmental pollutants), which may lead to a series of undesirable effects including jaundice, liver dysfunction and carcinogenesis. Several studies show that tolcapone, entacapone, licochalcone A, fructus psoraleae, amentoflavone displayed strong inhibitory effects on human UGT1A1 [1,18-20]. These findings provide a deep understanding of potential drug interactions between above mentioned drugs and UGT1A1 substrates via UGT1A1 inhibition, and will be helpful for guiding reasonable applications of above mentioned drugs and its related medical preparations in the future.
In our paper, products were denatured and spotted on positively charged nylon membranes instead of acrylamidemodified slides. After positively charged nylon membranes were baked at 100°C, some of the pyrimidine bases in the nucleic acid can be combined with the positive charges on the membranes. In the process of PCR products immobilization, no toxic substances were introduced, such as acrylamide monomer and TEMED. According to the three-dimensional polyacrylamide gel microarray technique, preparation of the acrylamide-modified slides is time-consuming [21]. Fluorescent labeled probes, acrylamide-modified primers and the use of Luxscan-10K/A (CapitalBio Company, China) are expensive. Based on the three-dimensional polyacrylamide gel microarray technique, dot blot hybridization, which is a safe, no toxicity and lower cost method, was used in our study. Although, dot blot hybridization analysis requires some labor for the detection of the allele-specific signals, this method is suitable for a large number of samples because a large number of samples can be blotted on one low-priced membrane and the costliest process in dot blot hybridization analysis is DIGlabeled probes.
In summary, we have successfully detected the UGT1A1 gene mutations in 15 Chinese patients with hyperbilirubinemia through dot blot hybridization method. This method will hold significant promise for applications in detection of UGT1A1 gene polymorphisms.
Acknowledgement
This work was supported by the Medical Science and Technology Development Foundation, Nanjing Department of Health (no. JQX14007) and Jiangsu Provincial Medical Youth Talent.
References
- Lv X, Wang XX, Hou J, Fang ZZ, Wu JJ, Cao YF, Liu SW, Ge GB, Yang L. Comparison of the inhibitory effects of tolcapone and entacapone against human UDP-glucuronosyltransferases. Toxicol Appl Pharmacol 2016; 301: 42-49.
- Guillemette C, Lévesque É, Rouleau M. Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin Pharmacol Therap 2014; 96: 324-339.
- Bosma PJ, Seppen J, Goldhoorn B, Bakker C, Oude Elferink RP, Chowdhury JR, Chowdhury NR, Jansen PL. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J Biol Chem 1994; 269: 17960-17964.
- Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. New England J Med 1995; 333: 1171-1175.
- Raijmakers MT, Jansen PL, Steegers EA, Peters WH. Association of human liver bilirubin UDP-glucuronyltransferase activity with a polymorphism in the promoter region of the UGT1A1 gene. J Hepatol 2000; 33: 348-351.
- Stingl JC, Bartels H, Viviani R, Lehmann ML, Brockmöller J. Relevance of UDP-glucuronosyltransferase polymorphisms for drug dosing: A quantitative systematic review. Pharmacol Therap 2014; 141: 92-116.
- Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenom 2003; 3: 136-158.
- Ginsberg G, Guyton K, Johns D, Schimek J, Angle K, Sonawane B. Genetic polymorphism in metabolism and host defense enzymes: Implications for human health risk assessment. Crit Rev Toxicol 2010; 40: 575-619.
- Matsui K, Maruo Y, Sato H, Takeuchi Y. Combined effect of regulatory polymorphisms on transcription of UGT1A1 as a cause of Gilbert syndrome. BMC Gastroenterol 2010; 10: 57.
- Fujiwara Y, Minani H. An overview of the recent progress in irinotecan pharmacogenetics. Pharmacogenomics 2010; 11: 391-406.
- Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: Dose matters. J Natl Cancer Institute 2007; 99: 1290-1295.
- Wenning LA, Petry AS, Kost JT, Jin B, Breidinger SA, DeLepeleire I, Carlini EJ, Young S, Rushmore T, Wagner F, Lunde NM, Bieberdorf F, Greenberg H, Stone JA, Wagner JA, Iwamoto M. Pharmacokinetics of raltegravir in individuals with UGT1A1 polymorphisms. Clin Pharmacol Therap 2009; 85: 623-627.
- Alaee E, Bazrafshan B, Azaminejad AR, Fouladinejad M, Shahbazi M. The association between prolonged jaundice and UGT1A1 gene polymorphism (G71R) in Gilbert's syndrome. J Clin Diagnos Res 2016; 10: 5-8.
- Song J, Sun M, Li J, Zhou D, Wu X. Three-dimensional polyacrylamide gel-based DNA microarray method effectively identifies UDP-glucuronosyltransferase 1A1 gene polymorphisms for the correct diagnosis of Gilbert's syndrome. Int J Mol Med 2016; 37: 575-580.
- Wong FL, Wang MK, Boo NY, Hamidah NH, Ainoon BO. Rapid detection of the UGT1A1 single nucleotide polymorphism G211A using real-time PCR with Taqman minor groove binder probes. J Clin Laboratory Analysis 2007; 21: 167-172.
- Hsieh TY, Shiu TY, Chu NF, Chao TY, Chu HC, Chang WK, Chao YC, Huang HH. Rapid molecular diagnosis of the Gilbert's syndrome-associated exon 1 mutation within the UGT1A1 gene. Genet Mol Res 2014; 13: 670-679.
- Shiu TY, Huang HH, Lin HH, Shih YL, Chu HC, Chang WK, Hsieh TY. Restriction fragment length polymorphism effectively identifies exon 1 mutation of UGT1A1 gene in patients with Gilbert's Syndrome. Liver Int 2015; 35: 2050-2056.
- Xin H, Qi XY, Wu JJ, Wang XX, Li Y, Hong JY, He W, Xu W, Ge GB, Yang L. Assessment of the inhibition potential of Licochalcone A against human UDP-glucuronosyltransferases. Food Chem Toxicol 2016; 90: 112-122.
- Wang XX, Lv X, Li SY, Hou J, Ning J, Wang JY, Cao YF, Ge GB, Guo B, Yang L. Identification and characterization of naturally occurring inhibitors against UDP-glucuronosyltransferase 1A1 in Fructus Psoraleae (Bu-gu-zhi). Toxicol Appl Pharmacol 2015; 289: 70-78.
- Lv X, Zhang JB, Wang XX, Hu WZ, Shi YS, Liu SW, Hao DC, Zhang WD, Ge GB, Hou J, Yang L. Amentoflavone is a potent broad-spectrum inhibitor of human UDP-glucuronosyltransferases. Chem Biol Interactions 2018; 284: 48-55.
- Rehman FN, Audeh M, Abrams ES, Hammond PW, Kenney M, Boles TC. Immobilization of acrylamide-modified oligonucleotides by co-polymerization. Nucleic Acids Res 1999; 27: 649-655.