ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2011) Volume 22, Issue 1
Molecular typing of an Acinetobacter baumannii outbreak based on inte-gron-gene cassette array-PCR
Bidur Prasad Chaulagain 1, 8, Dipendra Raj Pandeya2, Gyun-Yeol Ahn 1, Dong-Min Kim3, Jong-Hee Shin4, Joong-Ki Kook5, Sung-Heui Shin6, Dae-Soo Moon 1, Sook Jin Jang1, 7, *
1Department of Laboratory Medicine, Chosun University Medical School, Gwang-Ju, Korea.
2Department of Microbiology, Chonbuk National University Medical School, Jeonju, Korea
3Department of Internal Medicine, Chosun University Medical School, Gwang-Ju, Korea
4Department of Laboratory Medicine, Chonnam University Medical School, Gwang-ju, Korea
5Department of Oral Biochemistry, College of Dentistry, Chosun University, Gwang-ju, Korea
6Department of Microbiology, Chosun University Medical School, Gwang-Ju, Korea
7Department of Microbiology, Chosun University Medical School, Gwang-Ju, Korea
8Department of Plant Biology and Biotechnology, Faculty of Life Science, University of Copenhagen, Thorvaldsensvej 40, opg. 10. 1-1871 Frederiksberg, Denmark
- *Corresponding Author:
- Sook Jin Jang
Department of Laboratory Medicine/
Research Center for Resistant Cells
Chosun University Medical School,
588 Seoseok-dong, Dong-gu,
Gwang-Ju 501-717,
South Korea
Accepted date: September 18 2010
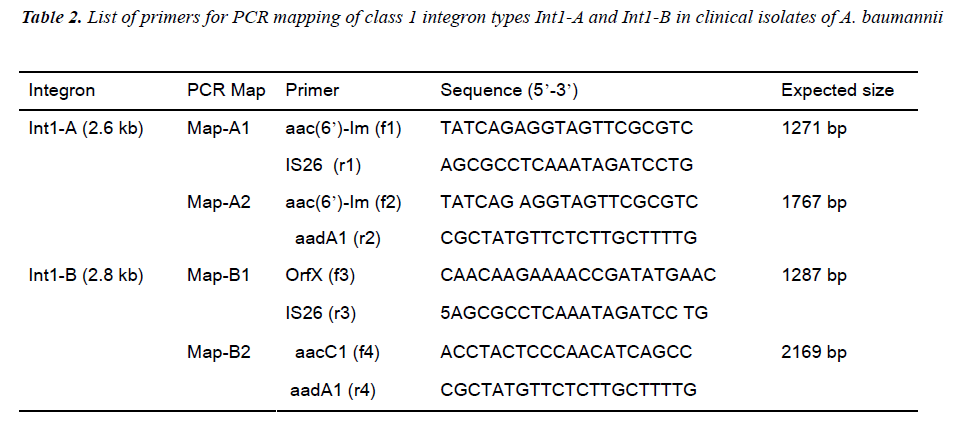
A total of 62 Acinetobacter baumannii isolates including 29 imipenem resistant, 23 imipenem susceptible and 10 environmental isolates were analyzed based on integron and integron gene cassette array (I-GCA) PCR and DNA sequences. Two types of class 1 integron were found based on their cassette arrays. The integron class 1 with a 2.6 kb size (Int1-A ) has the cas-sette order of aaC(6’)-Im followed by another gene cassette aadA1 disrupted by IS26 trans-posase, while the another class 1 integron with 2.8 kb size (Int1-B) has the gene cassette order of aacC1 followed by orfX, orfX’, IS 26 transposase and aadA1. PCR mapping detect the dis-tribution of such integron types within the isolates. Integron gene cassette array PCR (I-GCA PCR) typing gave 20 profiles while PFGE typing gave 18 profiles with 16 types and 2 sub-types. The major profile I of I-GCA PCR equivalent to profile A of PFGE was the dominant profile covering most of the clinical and environmental imipenem resistant isolates while the second largest profile XII of I-GCA PCR or profile M of PFGE was found in the imipenem susceptible isolates. Both integron types Int1-A and Int1-B were mostly restricted to imipenem resistant outbreak strains of the major profile I of I-GCA PCR or profile A of PFGE. I-GCA PCR based typing gave similar result to that of PFGE by sharing all the imipenem resistant strains of distinct profile I of I-GCA PCR in the profile A of PFGE which was responsible for the present A. baumannii outbreak. These results indicate that I-GCA PCR alone or coupled with other typing method would be a rapid tool for epidemiological study for bacteria that bear integron.
Keywords
Isolates, imipenem resistant, integron, epidemological
Introduction
There is worldwide occurrence of bacteremia caused by Acinetobacter baumannii (Ab). Now there are reports of increasing incidences of Acinetobacter baumannii out-breaks with carbapenem-resistance causing a variety of nosocomial infections in the intensive care units of hospi-tals [1,2] as well wound infections of the United States soldiers in war zones like middle east (Iraq) and South Asia (Afghanistan) [3]. A major contributing factor in the emergence of resistance of Acinetobacter species is the acquisition and transfer of antibiotic resistance genes on plasmids, transposons and integrons [4]. Among them integrons and gene cassettes are major contributor to the rapid increase in antibiotic resistance genes in clinical settings [5]. Integrons are found in bacterial genome spe-cially in the clinical bacteria and the β- and γ- protobacte-ria group [6]. Integron belongs to a class of genetic struc-tures that contain one or more gene cassettes with the fea-ture of antibiotics resistance. The integron has conserved 5’ segment (CS) and 3’ CS and upstream of 5’ CS of in-tegron possess the attI site, common promoter, Pant, and the opposite strand, the integrase gene, (intI) [7] and gene cassettes are embedded within the conserved region. Inte-grons are divided into different classes based on the se-quences of their integrase gene [8] and class 1 integron is the most common type followed by class 2 integron in A. baumannii and other gram negative bacteria.10
Many epidemiological studies have shown that integron could be a good genetic marker for the identification of epidemic strains [9] and it can be detected in various ways like 5’CS and 3’CS [10], integrase gene (intI) [11], inte-grase recombination site (attI) [6], gene cassette binding site (attC or 59-be) and integron promoters ( Pant) [12]. Gene cassettes in class 1 integron are common in Ab strains [13] and cassette PCR produces good results for electrophoretic band comparison [14].
Previously integrase gene PCR has been applied for the detection of different classes of integrons as well as to study epidemic behavior of Ab strains [15]. Despite the detail understanding of the molecular relationship be-tween antibiotic resistance gene cassettes and integrons in epidemic strains of clinical bacteria including Ab there are very few reports on the integron based molecular typing for epidemiological studies. At present we tried to under-stand the Acinetobacter outbreak based on integron gene cassette array polymerase chain reaction (I-GCA-PCR).
Material and Methods
Bacteria
An apparent hospital outbreak of imipenem-resistant A. baumannii (IRAB) occurred in the medical intensive care unit (MICU) and the surgical intensive care unit (SICU) of Chosun University hospital (CUH), a 707-bed tertiary care centre in Gwangju, South Korea, from January 2004 to December 2004. Clinical and environmental isolates collected during the epidemic period from MICU and SI-CU of CHU were analyzed for the present study. Fifty-two Acinetobacter baumannii clinical isolates, consisting 29 clinical-IRAB (C-IRAB), and 23 imipenem-suscep-tible A. baumannii (ISAB) control strains were studied for the molecular typing purpose (Table 1). Ten environ-mental IRAB (e-IRAB) isolates were also collected from different sources from the hospital to identify the source of IRAB contamination in the MICU and SICU.
Strain identification and antimicrobial susceptibility test was carried out by using the Vitek ID 32 GN system with Vitek AST-N017 kit (bioM?rieux, Durham, NC, USA) following the manufacturer’s instructions.
Preparation of genomic DNA (gDNA) material
Template DNA for present work was isolated from the cells of freshly cultured bacteria in LB agar medium, ei-ther with proteinase –K enzymatic extraction method [16] or from the bacterial cell lysate method [10]. DNA sam-ples then stored at − 20 °C in aliquots as PCR template material.
Molecular typing by PFGE
Macrorestriction analysis of chromosomal DNA of A. baumannii ApaI (New England Biolabs, Beverly, MA) was carried out by PFGE in a CHEF-DR III apparatus (BioRad Laboratories, Richmond, CA). Preparation of genomic DNA was done as described in ARPAC PFGE protocol http://www.hpa.org.uk/hpa/inter/arpac/a_typingprotocol). The restriction fragments were separated by a contour-clamped homogeneous electric field (CHEF-DRII system; BioRad, Munich, Germany) in 0.5×Tris–borate–EDTA buffer at 12°C and 200 V with pulse times of 5–13 s. DNA finger prints were interpreted as recommended by Tenover [17].
Polymerase chain Reaction
Integron and integrase PCR
For integrase gene PCR class 1 and class 2 integrase gene PCR primers and methods were applied as described by Koeleman [11] while for integrase class 3 PCR primer and method was used as mentioned by Senda [18]. The gen-eral scheme for integron PCR was followed as outlined byLevesque [10]. The integron common promoter (Pant) was detected with the set of primer and PCR conditions as described by Kim [12].
Sequencing of Integron
Amplified fragments obtained from class 1 integron PCR were purified by using PCR clean-up columns (Promega) and DNA was sequenced either directly by using the am-plification primers or cloned into pGEM®-T Easy (Pro-mega) vector according to the manufacturer's instructions. For cloned fragments the integron inserted plasmids were sequenced with primers based on the sequences of pGEM®-T Easy vector and by using internal primers whenever necessary for large DNA fragments. DNA se-quencing was performed at SolGent Sequencing Facility, Daejon, Korea by using an ABI Prism 377 (PE Biosys-tems, Foster City, CA). DNA sequences were used to search databases by using the National Center for Bio-technology Information BLAST web site (www.-ncbi.nlm.nih.gov:80/blast).
Integron-gene cassette array PCR (I-GCA PCR)
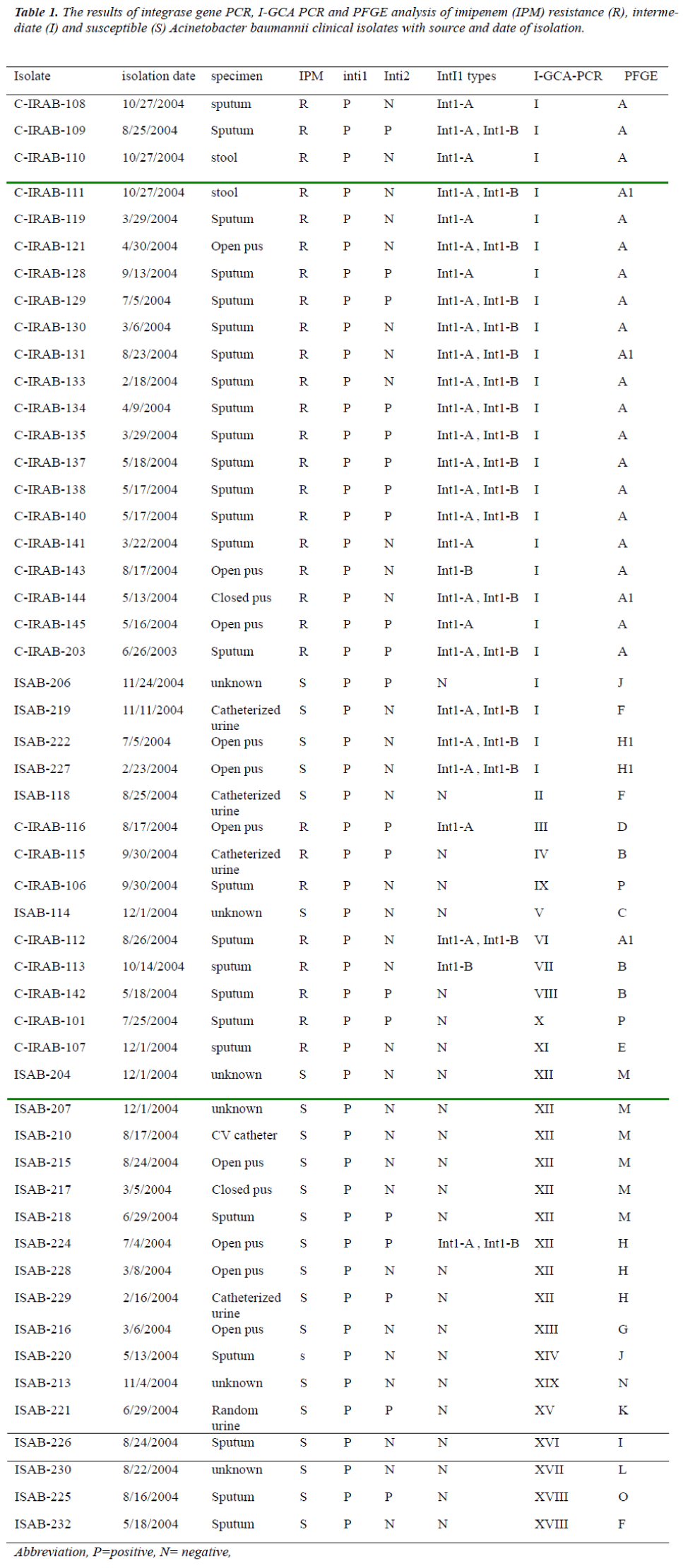
We used the class 1 integron sequences from our results to design different sets of primers for I-GCA- PCR using DNASTAR PrimerSelect program (DNASTAR v. 6.0). Primers sequences with different combinations of inte-gron components i) intI1 gene sequence as forward primer (Ac-intI1 F-5’-GCCGTAGAAGAACAGCAAGG-3’) and 59-be as reverse primer (Ac-59-be R - 5’- GTCTAACAATTCGTTCAA GC-3’), ii) attI as forward primer (TGTTTGATGTTATGGAGCAG) and Ac-59-be as reverse primer, iii) Pant as forward primer [12] and Ac-59-be as reverse primer and iv) Ac-59-be as both forward and reverse binding primers [6] were used for the I-GCA-PCR and all sets of primers gave the results applicable to fragment based typing purpose. The excellence for frag-ment analysis from the product was in the order of intI F-59-be R > attI F-59-be R > Pant F- 59-be R and the least performance was obtained from 59-be F-59-be R sets of primer for PCR fragment typing (Figure 1 and 2). The PCR products were run for electrophoresis in 0.9 % aga-rose gel in 0.5 X TBE buffer at 100V/cm. The gels were stained with 1mg/ml ethidium bromide solution [19] and photographed with Sanyo Digital camera and either printed or saved digitally in TIFF format for analysis. The selection of suitability of the primer sets were based on visual observation of the PCR amplicons on agarosoe gel and were not based on any statistical analyses.
Figure 1: Representative I-GCA PCR patterns with primer set Ac-Inti (forward) and Ac-59-be (reverse) from A. bau-mannii strains. Lanes: 1 and 17, molecular size marker; lanes 2 to 5, clinical imipenem resistant A. baumannii (C-IRAB) sporadic isolates; lanes 6 to 10 clinical imipenem resistant A. baumannii (C-IRAB) clonal isolates, lane 11, imipenem resistant A. baumannii environmental isolates (e-IRAB), 12 to 15, clinical imipenem susceptible A. bauman-nii isolates (ISAB), lane 16, negative control (PCR reaction mixture without DNA)..
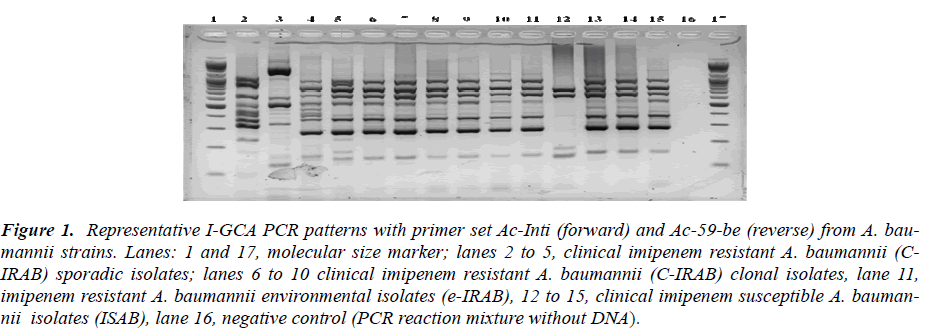
Figure 2: Representative I-GCA PCR patterns with primer set Ac-atti (forward) and Ac-59-be (reverse) from A. baumannii strains. lanes 1 to 15, clinical isolates of A. baumannii as described in Table 1, lane 16, negative control (PCR reaction mixture without DNA) and lane 17, molecular size marker. The top most row represents isolate name and lower row for profile types.
Analysis of the DNA patterns obtained by I-GCA PCR
I-GCA-PCR fragment patterns were compared by visual observation following the criteria set by Castaneda et al. [20]. Briefly, a single observer compared the pattern of each isolates against the rest and finger prints were con-sidered identical when all visible bands obtained by PCR had same number of bands with the same migration dis-tance irrespective of their band intensities. Patterns were considered different by the absence or presence of a sin-gle band. Sizes of DNA fragments amplified were deter-mined by comparison with the DNA marker. The analysis was performed blindly, by the assignment of a code to each isolate.
Integron screening in the isolates with PCR map
Based on the sequence information of class 1 integron was assayed by designing different sets of primer se-quences (Table 2). Two types of integrons, designated as Int1-A and Int1-B, PCR were revealed from the sequenc-ing result. PCR based integron types detection strategies were designed to detect the content and order of gene cas-settes of the integron in all of the test strains. Two PCR primer set for Int1-A assay was designed in an expecta-tion that the one amplicon product will be with the length of 1271 bp that covers some part of aaC(6’)-Im, and some parts of IS 26 inserted aadA1 gene (Map-A1) while another PCR was designed to amplify a product that cov-ers a part of aaC(6’)-Im in its proximal end and some parts of aadA1 in its distal end with an amplicon size of 1767 bp (Map-A2). Likewise for Int1-B screening the PCR Map-B1 was designed to cover some sequences of the orfX gene of the integron type Int1-B from the for-ward primer and reverse primer to cover the few se-quences of IS26 transposase to amplify a product of 1287 bp. Similarly the another PCR Map-B2 of Int1-B was designed to detect the integrity of gene cassettes aacC1 to aadA1 covering all the DNA sequences between two proximities with a amplicon size of 2169 bp out of a 2875 bp of the full sequence of Int1-B integron in this study (Figure 1 and 2).
Results
Genotyping of A. baumannii by pulsed-field gel electro-phoresis (PFGE)
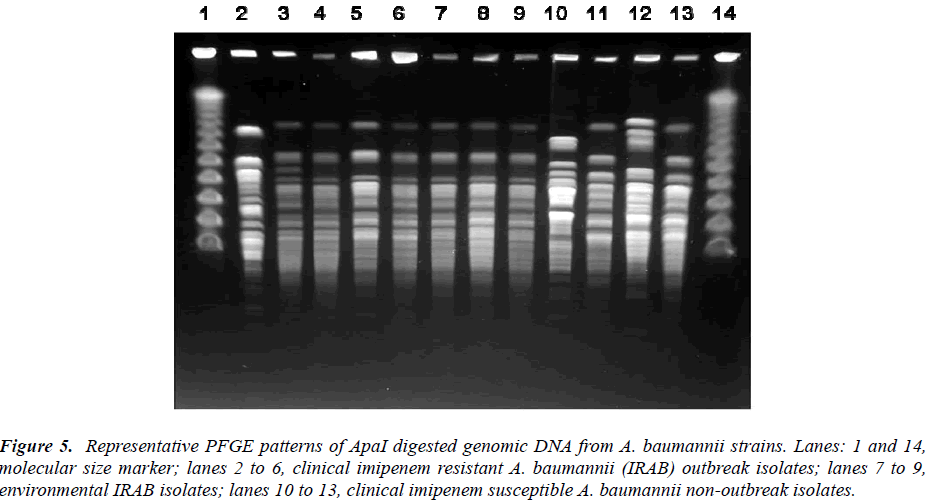
The representative PFGE patterns are shown in Figure 5 and Table 1. The typing gave 16 types and two subtypes. Among the 29 C-IRAB isolates the major outbreak strain was represented by PFGE profile A (23/29, 79.31%) among the 52 clinical isolates sharing 44.23% (23/52) of the total isolates. Other minor clonal types were type M (6/52, 11.54%), type H (5/52, 9.62%), type F (3/52, 5.77%)and J (2/52, 3.85%) all representing the ISAB strains.
Figure 5: Representative PFGE patterns of ApaI digested genomic DNA from A. baumannii strains. Lanes: 1 and 14, molecular size marker; lanes 2 to 6, clinical imipenem resistant A. baumannii (IRAB) outbreak isolates; lanes 7 to 9, environmental IRAB isolates; lanes 10 to 13, clinical imipenem susceptible A. baumannii non-outbreak isolates.
Integrase PCR and Integron types
All the detected Ab isolates were positive for integron based on integrase gene PCR for class 1 integron as well as integron promoter (Pant) PCR. Integron class 2 was rep-resented by 38.46% (20/52) isolates when assayed with integrase gene (intI2) PCR while class 3 integron was absent in the tested isolates (Table 1). Since all the tested isolates were positive to class 1 integron we further de-signed I-GCA PCR based on the DNA sequence informa-tion of the class 1 integron.
All the detected Ab isolates were positive for integron based on integrase gene PCR for class 1 integron as well as integron promoter (Pant) PCR. Integron class 2 was rep-resented by 38.46% (20/52) isolates when assayed with integrase gene (intI2) PCR while class 3 integron was absent in the tested isolates (Table 1). Since all the tested isolates were positive to class 1 integron we further de-signed I-GCA PCR based on the DNA sequence informa-tion of the class 1 integron.
Integron PCR mapping
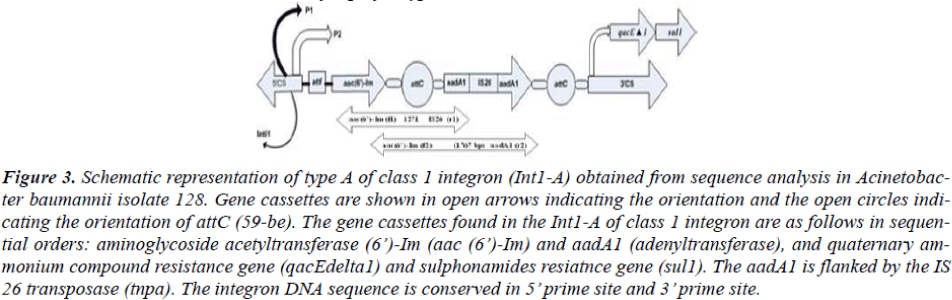
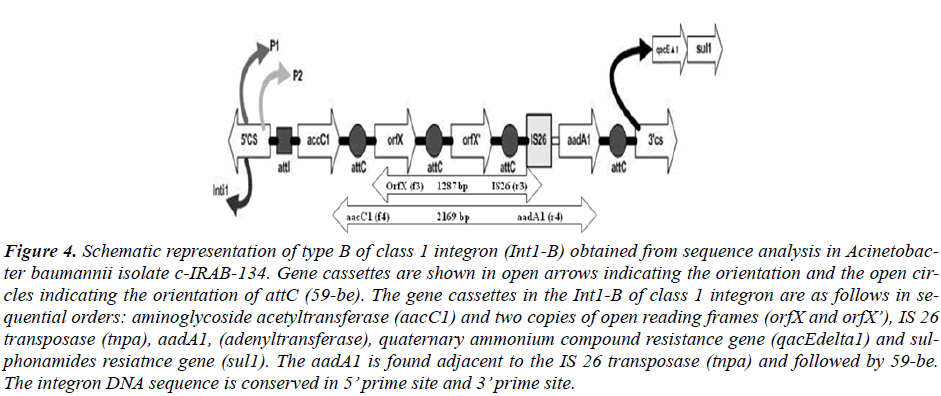
Based on the sequence information about the orders of inserted gene cassettes integron PCR mapping was done with two sets of primers for each integron types. (Table 2, Figure 3 and 4). The distribution of Int1-A and Int1-B tested in all of the isolates are described in Table 1.
Figure 3: Schematic representation of type A of class 1 integron (Int1-A) obtained from sequence analysis in Acinetobac-ter baumannii isolate 128. Gene cassettes are shown in open arrows indicating the orientation and the open circles indi-cating the orientation of attC (59-be). The gene cassettes found in the Int1-A of class 1 integron are as follows in sequen-tial orders: aminoglycoside acetyltransferase (6’)-Im (aac (6’)-Im) and aadA1 (adenyltransferase), and quaternary am-monium compound resistance gene (qacEdelta1) and sulphonamides resiatnce gene (sul1). The aadA1 is flanked by the IS 26 transposase (tnpa). The integron DNA sequence is conserved in 5’ prime site and 3’ prime site.
Figure 4: Schematic representation of type B of class 1 integron (Int1-B) obtained from sequence analysis in Acinetobac-ter baumannii isolate c-IRAB-134. Gene cassettes are shown in open arrows indicating the orientation and the open cir-cles indicating the orientation of attC (59-be). The gene cassettes in the Int1-B of class 1 integron are as follows in se-quential orders: aminoglycoside acetyltransferase (aacC1) and two copies of open reading frames (orfX and orfX’), IS 26 transposase (tnpa), aadA1, (adenyltransferase), quaternary ammonium compound resistance gene (qacEdelta1) and sul-phonamides resiatnce gene (sul1). The aadA1 is found adjacent to the IS 26 transposase (tnpa) and followed by 59-be. The integron DNA sequence is conserved in 5’ prime site and 3’ prime site.
The type Int1-A has the gene cassette aaC (6’)-Im in the most proximal side nearest to the 5’CS while the type Int1-B harbored aacC1 cassette nearest to the 5’CS. The nearest cassettes to the promoter are considered to be best expressed among the cassettes in the integron in compari-son to other inserted cassettes which are distant from the Pant [7].
I-GCA-PCR and PFGE profile distribution among iso-lates
Fifty three clinical isolates were typed by I-GCA-PCR. All the isolates were grouped into 20 types based on the PCR fragment migration in the agarose gel electrophore-sis (Table 1, Figure 1and 2). The common five PCR frag-ments of profile I of I-GCA-PCR were found in 48.08 percent (25 of the 52) of clinical isolates followed by 6 bands type in profile XII which shared more than 17.31 percent (9 of 52) isolates and the profile XVIII were found in about 3.85 percent (2 of the 52) and rest of the isolates grouped under 16 sporadic I-GCA-PCR patterns were distributed only in 30.77 percent (16 of 52) of total isolates which were mostly imipenem susceptible.
The I-GCA-PCR grouped the 52 isolates into 20 types while the Apa digested PFGE also grouped them into 18 types (with 16 types and 2 subtypes). The I-GCA-PCR showed in total 3 clonal groups while the PFGE has given 7 clonal groups. Those isolates of I-GCA-PCR major clonal type I that were not included in PFGE clonal type A were distributed in other clonal group of PFGE such as two isolates in profile H and its subtype H1. Interest-ingly the I-GCA-PCR clonal type XII included 9 isolates of which 6 isolates were from PFGE clonal group M and 3 isolates from clonal group H. But none of the isolates of I-GCA-PCR major clonal type I, XII and XVIII were dis-tributed in the sporadic profiles of PFGE with exception for one isolate of minor clone XVIII distributed in PFGE clone Q.
Distribution of class 1 integron types Int1-A and Int1-B among the Ab Isolates
Int1-A was distributed in 50.00 % (26 of 52) of the clini-cal isolates with clustering in C-IRAB (22/29, 75.86%) while Int1-B was distributed in 40.38% (21/52) of total clinical isolates and 72.41% (21/29) of C-IRAB isolates. Both the integron types Int1-A Int1-B were mostly dis-tributed in profile I of I-GCA PCR and the distribution of types Int1-A Int1-B in other profile is sporadic. Isolates carrying both types Int1-A Int1-B types were 36.54% (19 out of 52) and mostly concentrated in profile I (89.47 %, 17/19).
Discussion
At present class 1 integron was observed in all of the tested isolates. The prevalence of integrase class 1 in A. baumannii isolates in our hospital in Korea was similar to the previous report of the period of 1998 in another hospi-tal located in another city for Ab and Acinetobacter DNA group 13TU isolates [21]. This shows Korean Ab strains have unique tendency to bear integron in all strains in contrary to other studies where integrons were found in outbreak strains as a marker [11, 22].
When we did sequence analysis and searched against NCBI BLAST class 1 integron show two different cassette arrays designated here as Int1-A and Int1-B. Both inte-gron types were detected in the C-IRAB outbreak strains being distributed in the profile I of I-GCA-PCR and pro-file A of PFGE with rare distribution in other profiles. Although very rare such occurrence of integron (types Int1-A and Int1-B) in other sporadic profile is also an widely observed and expected phenomenon in light of horizontal gene transfer since intra-strains to interspecies transfer of integrons is natural in bacteria sharing the common habitat [13,23].
The sequencing of the class 1 integron gave two types of gene cassettes order. The class 1 integron with type int1-A with a 2.6 kb size has the cassette order aaC (6’)-Im fol-lowed by aadA1 disrupted by IS26 transposase, the order being extended from the 5’CS region to the 3’CS region. Likewise, the type Int1-B has the gene cassette order of aacC1 followed by orfX, orfX’, IS 26 transposase and aadA1. Both the features are unique to integron class 1 and confined to our hospital and another neighboring hospital in Gwangju city of Korea [24]. In integron gen-erally a new mobile gene cassette attaches at the attI site [25] but recombination may also occur between two 59-be’s of mobile gene cassettes [25]. In present case the aadA1-IS-26-aadA1 gene cassette of integron type Int1-A and IS 26- aadA1 of integron type Int1-B are found at-tached at the vicinity of 3’ CS of integron instead of at-taching to the attI site near to 5’ CS region which depicts a rare case of integron bearing by Acinetobacter at present case. Such unusual insertion of integron gene cassettes possibly may have contributed for the imipenem resis-tance in those strains which needs further study. The beta-lactamase PCR assay did not show any beta-lactamase gene either in the integron or on chromosome in the tested strains (data not shown).
We tried to develop the I-GCA-PCR typing method and evaluated the result comparing with PFGE which is taken as a gold standard for the molecular typing purpose. A total of 52 clinical and 10 environmental isolates were typed by I-GCA-PCR (Table 1). All the typed isolates were grouped into 19 I-GCA-PCR profiles and the ge-nomic DNA of ApaI restriction digested PFGE gave 16 types and 2 subtypes. Of the 52 clinical isolates 25 (48.08%) isolates fell in the major profile I of I-GCA-PCR and the PFGE profile A included 22 (41.50 %) iso-lates as the major profile. Another I-GCA-PCR profile XII included 17.31% (9/52) of isolates while including 6 out of 9 isolates from the XII profile in the PFGE profile M. The remained clonal I-GCA-PCR profile XVIII in-cluded 2 isolates. All the environmental strains isolated from different objects of hospital settings and from the hands of HCWs were also typed as profile I of I-GCA-PCR and profile A of PFGE and imipenem resistant simi-lar to C-IRAB.
The genotyping shows the clear feature of IRAB outbreak with a genetic distinction between the C-IRAB and ISAB isolates.. The major clone I of I-GCA-PCR or A of PFGE was the most prevalent, found in most of the imipenem resistant isolates where the second genotypic profile XII of I-GCA-PCR or clone M and H of PFGE were found in imipenem susceptible isolates.
Concordance of typing methods is a good principle to validate the identification of distinct clonal group in dif-ferent typing techniques since natural groups must be identical or very similar when tested by methods that measure independent markers [26]. While comparing mo-lecular typing systems for epidemiological studies the distribution of strains sharing the common features has significance. The matching of distribution of C-IRAB isolates in the major clone I of I-GCA-PCR profile with profile A of PFGE shows the applicability of I-GCA-PCR for the genotyping purpose in epidemiological study. At present like PFGE, the I-GCA PCR typing system in-cluded all the C-IRAB outbreak clones characterized by imipenem resistance in the major profile and the imipe-nem susceptible isolates in other minor profiles or in dis-tinct sporadic profiles. I-GCA PCR gave 20 strain profiles very similar to PFGE typing with 18 profiles. The major clones of both typing systems shared all the isolates with common features showing the concordance of I-GCA-PCR typing with PFGE. This result suggests the applica-tion of I-GCA-PCR for epidemiological studies in micro-organisms bearing integron.
In summary, I-GCA-PCR and integron PCR mapping helped us to speculate that present outbreak was spread from a single clone I of I-GCA-PCR equivalent to clone A of PFGE. Thus integron based typing system like I-GCA-PCR can work as a rapid tool to address bacterial out-break that bears integron in its genome. It is a case of unique strains with unique integron types confined to our hospital in Korea.
Acknowledgement
This work was supported by the National Research Foun-dation of Korea (NRF) grant funded by the Korea gov-ernment (MEST) through the Research center of Resistant Cells (R13-2003-009)
References
- Gonzalez GK, Sossa H, Bello M, Dominguez S, Ze-melman R. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol. Lett 1998; 161: 125-128.
- Zarrilli R, Crispino M, BagattIni M, Barretta E, Di Po-polo A, Triassi M, et al. Molecular epidemiology of se-quential outbreaks of Acinetobacter baumannii in an in-tensive care unit shows the emergence of carbapenem resistance. J Clin Microbiol 2004; 42: 946-953.
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii, Antimicrob. Agents Chemother. 2007; 51: 3471-3484.
- Nemec A, Dijkshoorn L, Van der Reijden TJ. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J. Med. Microbiol. 2004; 53: 147-153.
- Hall RM, Stokes HW. Integrons: novel DNA elements wake capture genes by site-specific recombination. Ge-netica 1993; 90: 115-132.
- Gillings MR, Holley MP, Stokes HW, Holmes AJ. In-tegrons in Xanthomonas: A source of species genome diversity. Proc. Natl. Acad. Sci. USA. 2005; 102: 4419-4424.
- Hall RM, Collis CM. Mobile gene cassettes and inte-grons: capture and spread of genes by site-specific re-combination. Mol. Microbiol. 1995; 15: 593-600.
- Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acineto-bacter baumannii. FEMS Microbiol. Lett. 2006; 258: 72-77.
- Valenzuela JK, Thomas L, Patridge SR, Van der Rei-jden T, Dijkshoorn L, Iredell J. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii.J. Clin. Microbiol. 2007; 45: 453-460.
- Levesque C, Piche L, Chantal L, Roy PH. PCR map-ping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents. Chemother. 1995; 39: 185-191.
- Koeleman JGM, Van der Bjil MW, Stoof J, Vanderbroucke-Grauis CMJE, Savelkoul PHM. Antibi-otic resistance is a major risk factor for epidemic be-haviour of Acinetobacter baumannii. Infect. Control Hosp. Epidimiol. 2001; 22: 284-288.
- Kim YH, Cho K, Yun SH, Kim JY, Kwon KH, Yoo JS, Kim SI. Analysis of aromatic catabolic pathways in Pseudomonas putida KT2440 by combined proteomic approach: 2-DE/MS and cleavable ICATabalysis. Pro-teomics 2006; 6: 1301-1318.
- Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti-Bragadin C. et al. Molecular charac-terization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Ital-ian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob. Agents Chemother. 2002; 46: 3665-3668.
- Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding specific gene-integration functions: integrons. Mol. Microbiol. 1989; 3: 1669-1683.
- Turton JF, Kaufmann ME, Warner M, Coelho JM, Dijkshoorn L, van der Reijden T. et al. A prevalent, multi-resistant, clone of Acinetobacter baumannii in South East England. J. Hosp. Infect. 2004: 58:170-179.
- Sambrook J, Russell DW. Extraction and purification of plasmid and screening of bacterial colonies by hybridi-zation, p. 1.40-1.92. In J. Argentine, N. Irwin, K. A. Jassen, S. C. M. Zierler, N. Mclnerny, D. Brown, and S. Schaefer (ed.), Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. 2001.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromo-somal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 1995; 33: 2233-2239.
- Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K. et al. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resis-tant to broad-spectrum β-lactams, including carbap-enems. Antimicrob. Agents Chemother. 1996; 40: 349-353
- Sambrook J, Fritsch EF, Maniatis T. Molecular Clon-ing: a Laboratory Manual, 2nd edn. Cold Spring Har-bor, NY: Cold Spring Harbor Laboratory. 1989.
- Castaneda M, Sanchez J, Moreno S, Nunez, C, Espin, G. The global regulators GacA and sigma (S) form part of a cascade that controls alginate production in Azoto-bacter vinelandii. J. Bacteriol. 2001; 183: 6787-6793.
- Oh EJ, Lee S, Park YJ, Park JJ, Park K, Kim SI. et al. Prevalence of metallo-ß-lactamase among Pseudomo-nas aeruginosa in Korean University hospital and comparison of screening methods for detecting metallo-ß-lactamase. J. Microbiol. Method. 2003; 54: 411-418.
- Kraniotaki E, Manganelli R, Platsouka E, Grossato A, Paniara O, Palu G. Molecular investigation of an out-break of multi drug resistant Acinetobacter baumannii, with characterization of class 1 integrons. J. Antim-icrob. Agents 2006; 28: 193-199.
- Preston KE, Radomski CC, Venezia RA. The cassettes and 3' conserved segment of an integron from Kleb-siella oxytocaplasmid pACM1. Plasmid 1999; 42: 104-114.
- Ok YJ, Jang SJ, Li XM, Park G, Yong DE, Kang JO. et al. Prevalence and spread of integron-IS26 in imipenem-resistant Acinetobacter baumannii clinical isolates in South Korea. Int. J. Antimicrob. Agents 2009; 34:609-611.
- Labbate M, Boucher Y, Joss MJ, Michael CA, Gillings MR, Stokes HW. Use of chromosomal integron arrays as a phylogenetic typing system for Vibrio cholerae pandemic strains. Microbiol. 2007; 153: 1488-1498.
- Struelens MJ, the Members of the European Study Group on Epidemiological Markers (ESGEM), of the European Society for Clinical Microbiology and Infec-tious Diseases (ESCMID). Consensus guidelines for appropriate use and evaluation of microbial epidemi-ologic typing systems. Clin Microbiol Infect. 1996; 2: 2-11.