ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 3
Occurrence of blaCTXM-2, blaSHV, blaTEM genes in ESBL-producing bacteria from retail sausages in Kampar, Malaysia.
Li-Yen She, Lih-Shin Tew and Choy-Hoong Chew*
Department of Biomedical Science, Faculty of Science, Universiti Tunku Abdul Rahman, Jalan Universiti, Bandar Barat, 31900 Kampar, Perak, Malaysia
- *Corresponding Author:
- Choy-Hoong Chew
Department of Biomedical Science
Faculty of Science
Universiti Tunku Abdul Rahman
Jalan Universiti, Bandar Barat
31900 Kampar, Perak Malaysia.
Accepted March 14 2014
Overuse of antibiotics in the prevention and treatment of diseases among humans and livestock has led to the emergence of multi-antibiotics resistant bacteria worldwide. This includes bacteria which produce the enzymes extended-spectrum β-lactamases (ESBLs). This study reports the presence of β-lactamases-producing bacteria (35.7%) and ESBL-producing bacteria (21.4%) from retail sausages in Kampar, Malaysia and the detection of blaCTXM-2 (60%), blaTEM (40%) and blaSHV (20%) genes, but not blaCTXM-1 and blaCTXM-9 genes in these isolates. The bacteria harboring blaCTXM-2 and blaTEM genes were identified as Pseudomonas pneumotropica, whereas the bacteria harboring only blaSHV gene was identified as Klebsiella pneumoniae subsp. pneumoniae. This is the first report of blaCTXM-2 in food source in Malaysia.
Keywords
antibiotic resistant ESBL-producing bacteria, sausages, blaCTXM-2, blaSHV, blaTEM
Introduction
The overuse of antibiotics in human medicine, animals and agriculture has led to selective pressures in the adverse environment and the emergence of antibiotics resistant genes among bacteria. This has led to a great deal of attention being paid to the emergence of antibiotic resistant bacteria in both human and animal populations. As antibiotic resistant bacteria can lead to the risks of spread of resistant strains among humans and animals, this impacts morbidity and mortality from chronic diseases and increases the cost of therapy [1].
One of the current important antibiotic resistance mechanisms is the plasmid-mediated production of β- lactamases. This enzyme is able to inactivate the β-lactam antibiotics by hydrolyzing the β-lactam ring [2]. However, the prolonged exposure of bacteria to a variety of β- lactams antibiotics has led to continuous mutation and production of β-lactamases and expanding their activity towards the newly developed β-lactam antibiotics [3]. These enzymes are known as Extended-Spectrum β- Lactamases (ESBLs) and are most frequently reported as TEM, SHV and CTX-M types [1]. ESBLs confer broad resistance to penicillins, monobactams (aztreonam) and up to third and fourth generations of cephalosporins (with the exception of cephamycins and carbapenems) [4]. However, they are inhibited by sulbactams, tazobactams and clavulanic acid, a feature that is used as a standard for β-lactamases classification and for diagnostic ESBL detection purposes [2].
Recently, the recovery of ESBL-producing bacteria from different types of food products and animals was reported in some studies [5-7]. These reports have raised questions about the possible interplay of animal- and food-related reservoirs in the spread of ESBL- producing microorganisms. Hence, this study was conducted to isolate the ESBL-producing bacteria from retail sausages of poultry source, in addition to identify the antibiotic resistance and gene determinants.
Materials and Methods
Sample collection, isolation and identification
All the sausages were collected from a hypermarket and cafeteria of the university in Kampar, Perak, Malaysia. Each of the sausages was cut into small pieces to approximately two grams and cultured into five replicates of Luria-Bertani (LB) enrichment broths (Laboratories CONDA, Spain) respectively on the day of arrival.
The culture broths were then incubated at 37oC for 16- to 18-hours and a loopful of the enrichment broth was streaked onto MacConkey (MAC) agar (Oxoid, England) plates for the isolation of Gram-negative bacteria. The agar plates were then incubated aerobically at 37oC for 24-hours. Any of the gram-negative bacteria isolated from retail sausages were then subjected to a range of general biochemical tests and identified using the API 20E bacterial identification kit (bioMerieux®, France) in accordance to the manufacturer’s specifications.
Screening and confirmation of ESBL-producing bacteria
Initial screening test for ESBL-producing bacteria was performed by using Kirby-Bauer disc diffusion antibiotic susceptibility test. A single colony of pure culture from the overnight MacConkey agar plate was inoculated into 5 mL of Trpytic Soy Broth (TSB) (Merck, Germany) and incubated at 37oC overnight. The turbidity of the inoculums in the TSB was adjusted to 0.5 McFarland standard and the plates were screened with five types of antibiotics, which included aztreonam (ATM-30), cefotaxime (CTX-30) (BD BBLTM, United States), cefpodoxime (CPD-10), ceftriazone (CRO-30) and ceftazidime (CAZ- 30) (Oxoid, England), based on the Clinical and Laboratory Standards Institute (CLSI) standard [8]. Any isolate that showed resistance to any of the antibiotic in initial screening test was subjected to double disc synergy test for ESBL production.
In this method, a disc containing amoxicillin-clavulanic acid (AMC-30) (Oxoid, England) was placed at the center of Mueller-Hinton agar. Discs containing ceftazidime (CAZ-30) and cefotaxime (CTX-30) (BD BBLTM, United States) were applied 15 mm apart from the disc at the center, which was the amoxicillin-clavulanic acid (AMC-30) in accordance to CLSI protocols. After overnight incubation at 37oC, ESBL enzyme production by the bacteria isolated would be indicated by a keyhole zone phenomenon towards the amoxicillin-clavulanic acid (AMC-30) disc. Phenotypic confirmatory test for ESBL production was carried out using combination disc method. An increase of ≥ 5 mm in a zone diameter for either antimicrobial agent (cefotaxime (CTX-30) or ceftazidime (CAZ-30)) tested in combination with clavulanic acid versus the zone diameter of the agent when tested alone after incubation at 37oC for 16- to 18-hours was considered positive for ESBL production [8].
Antibiotic susceptibility testing
In addition, the bacteria were also subjected to further Kirby-Bauer antibiotic susceptibility tests using additional nine types of antibiotics, which were ampicillin (AMP- 10), penicillin (P-1), oxacillin (OX-1), piperacillin (PRL- 75), imipenem (IMP-10), meropenem (MEM-10), norfloxacin (NOR-10) (Oxoid, England), gentamicin (CN- 10), and tetracycline (TE-30) (BD BBLTM, United States).
Total DNA extraction and Polymerase chain reaction
Total DNA was extracted using fast-boil method as highlighted in Kor et al., 2013 [9]. Using established primers of blaCTXM-1, blaCTXM-2, blaCTXM-9, blaTEM and blaSHV [10- 11], PCR amplifications were carried out using PCR reagents from Invitrogen (USA) and established conditions as listed by the authors. The amplified products were subjected to 1.5% (w/v) agarose gel electrophoresis, stained with ethidium bromide before visualization under UV light using the UV transilluminator (Syngene, United Kingdom).
Results
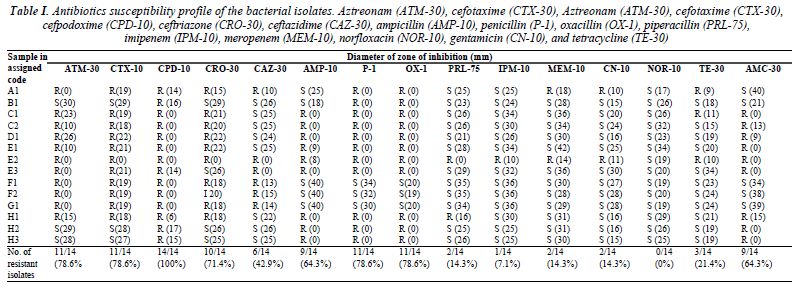
A total number of 14 bacterial strains with different morphologies were isolated from eight different types of sausages. The zone of inhibition in diameter to the nearest millimeter was measured for each antibiotic tested and summarized in Table I. In this study, 11 (78.6%) out of the 14 bacterial isolates were resistant to the aztreonam, 11 (78.6%) bacterial isolates showed resistance to cefotaxime, all of the bacteria (100%) were resistant to cefpodoxime, while 10 (71.4%) of the bacteria were resistant to ceftriazone. Majority of the bacterial isolates were susceptible to ceftazidime as only six (42.9%) were resistant to ceftazidime.
Table I. Antibiotics susceptibility profile of the bacterial isolates. Aztreonam (ATM-30), cefotaxime (CTX-30), Aztreonam (ATM-30), cefotaxime (CTX-30), cefpodoxime (CPD-10), ceftriazone (CRO-30), ceftazidime (CAZ-30), ampicillin (AMP-10), penicillin (P-1), oxacillin (OX-1), piperacillin (PRL-75), imipenem (IPM-10), meropenem (MEM-10), norfloxacin (NOR-10), gentamicin (CN-10), and tetracycline (TE-30)
Besides, high resistance was also observed towards ampicillin (64.3%), penicillin and oxacillin (78.6%), while the isolates were more susceptible towards piperacillin and the carbapenem and tetracycline group of antibiotics. All (100%) the bacteria were susceptible to norfloxacin.
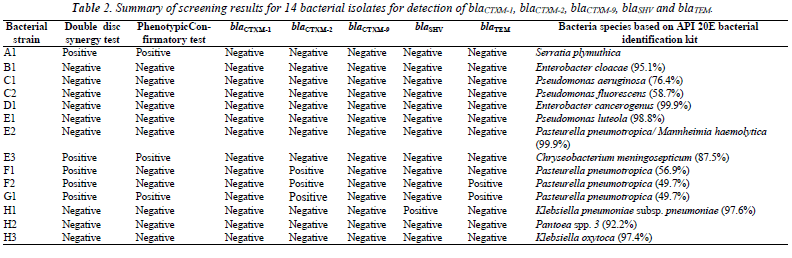
As shown in Table II, five (35.7%) bacterial isolates (A1, E3, F1, F2 and G1) showed the keyhole zone phenomenon on the Mueller-Hinton agar, and are possible ESBLproducers. In phenotypic confirmatory test, only three isolates (A1, E3 and G1) showed ≥ 5 mm increase in the zone diameter around the disc with clavulanic acid over the disc with ceftazidime alone.
None of the bacterial isolate harbored any blaCTXM-1 and blaCTXM-9 genes, but there was a high prevalence of blaCTXM-2 among the bacterial isolates from sausages. Three out of the five possible ESBL-producers (F1, F2 and G1) were screened positive for the presence of blaCTXM-2. Both F2 and G1 also co-harbored blaTEM. and were identified to be Pasteurella pneumotropica although only the latter showed positive in phenotypic confirmatory test. Interestingly, the isolate H1 which was resistant towards nine of the antibiotics including cephalosporin and augmentin, but showed negative results for double disc synergy and phenotypic confirmatory tests, was the only isolate which was tested positive for blaSHV and was identified to be Klebsiella pneumoniae subsp. pneumoniae. Both isolates A1 and E3 showed positive results for both the double disc synergy and phenotypic confirmatory tests, but they did not harbor any of the ESBL gene tested.
Discussion
As stated in CLSI standard, the use of more than one type of antimicrobial agent for screening purpose can improve the sensitivity of ESBL production. In year 1988, double disc synergy test was described by Jarlier and others to detect the production of ESBL enzyme in K. pneumoniae and E. coli isolates. This keyhole zone phenomenon is also known as “lens of inhibition”, which is an extension in the zone of inhibition surrounding the peripheral discs towards the disc placed at the centre caused by the synergy activity between the clavulanic acid and cefotaxime or ceftazidime.
However, the guideline recommendations for phenotypic confirmatory test by CLSI are restricted to a few types of bacteria, which are K. pneumoniae, K. oxytoca, E. coli and P. mirabilis [8]. In agreement with some researchers, CLSI standard guidelines should be extended to other bacteria genus and species as long as they do not produce any false- negative or positive results [12-14]. Indeed, it has been shown that any bacteria which produce KPCbeta lactamases or hyperproduction of K1-beta lactamases may cause false-positive results, whereas isolates that produced high levels of AmpC enzymes may cause falsenegative results [12]. Hence, these bacteria will interfere with phenotypic tests interpretation of ESBL-producers. As observed in Table I and Table II, nine of the isolates proved to be augmentin resistant although they were resistant to at least one cephalosporin or showed positive keyhole phenomenon. These could indicate carriage of AmpC genes which leads to augmentin resistance, and the false negative results in the isolates. However, this remains to be investigated.
Both F2 and G1 which co-harbored blaCTXM-2 and blaTEM were identified to be Pasteurella pneumotropica. P. pneumotropica is part of the commensal oropharyngeal floras of many animals, including dogs and cats, and is usually considered a rare but opportunistic human pathogen [15]. Interestingly, this organism is not commonly reported from food source.
Although there have been reports of blaCTXM from patient isolates in Malaysia [16], our study here is the first report of the high prevalence of blaCTXM-2 gene amongst possible ESBL-producers isolated from food source in Malaysia. As highlighted by Liebana et al. [17], blaCTXM-1, blaCTXM-14, and blaCMY-2 are usually the most frequent genes identified in Escherichia coli and Salmonella from food animals. In most countries like United Kingdom, South India and Iran, blaCTXM-1 was the most predominant gene found in ESBL isolates [10, 18-19]. In a study by Luvsansharav et al. [20], blaCTXM-9 was predominant in ESBL-producing Enterobacteriaceae among the fecal carriage in rural Thai communities followed by the blaCTXM-1. Likewise, blaCTXM-2 has been detected in Israel, Japan, and most South American countries [21-23].
Interestingly, the isolate H1 was identified to be Klebsiella pneumoniae which was the only bacterium that harbored the ESBL gene categorized under Enterobacteriaceae family in this study. This is rather unexpected because high prevalence of Enterobacteriaceae are usually isolated and reported from animal food. Indeed, several types of Enterobacteriaceae that harbored ESBL genes were isolated from ready-to-eatfruits in Nigeria such as Klebsiella oxytoca (31.7%), Klebsiella pneumoniae (22.0%), Enterobacter gergoviae (13.4%), Pantoea agglomerans (3.7%) and so on [24]. Besides, there was also report of a large nosocomial outbreak through the food chain in an acute care hospital in Barcelona, Spain, for the dissemination of blaSHV-1 and blaCTX-M-15 producing K. pneumoniae [25].
In this study, both Serratia plymuthica (isolate A1) and Chryseobacterium meningosepticum (isolate E3) showed positive results for both the double disc synergy and phenotypic confirmatory tests, but they did not harbor any ESBL gene tested here. This may due to the reason where both these bacteria harbored other types of ESBL genes that were not tested in the study, which included blaAmpC gene and blaOXA gene [26-27]. This is possible as S. plymuthica was shown to be resistant to oxacillin, while C. meningosepticum was resistant to both the ampicillin and oxacillin.
In conclusion, this is the second report of ESBL-genes from poultry food source after blaSHV was discovered in sushi [11]. It is paramount that control measures are to be implemented to minimize public health risk through the consumption of contaminated food. Hence, efforts should be directed to increase awareness and hygienic practices during the food processing and postharvest procedures.
References
- Amador P, Fernandes R, Brito L, Prudencio C. Antibioticresistance in Enterobacteriaceae isolated from Portugesedeli meat. Journal of Food Safety 2011; 31: 1-20.
- Geser N, Stephan R, Hächler H. Occurence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Veterinary Research 2012; 8: 1-9.
- Shah AA, Hasan F, Ahmed S, Hameed A. Characteristics, epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extended-spectrum β- lactamases. Research in Microbiology 2004; 155: 409- 421.
- Adenaike O, Olonitola OS, AmehJB,Whong CMZ. Incidence of extended spectrum β-lactamase producing bacteria and multidrug resistance strains from processed meat "suya" sold in a University Community. The International Journal of Engineering and Science 2013; 2: 1-6.
- Angulo FJ, Nargund VN, Chiller TC. Evidence of an association between use of antimicrobial agents in food animals and antimicrobial resistance among bacteria isolated from humans and the human health consequences of such resistance.Journal of Veterinary Medicine B, Infectious Diseases and Veterinary Public Health 2004; 51(8-9):374-9.
- Buchholz U, Bernard H, Werber D et al. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. New England Journal of Medicine 2011; 365: 1763-1770.
- Bergenholtz RD, Jorgensen MS, Hansen LH, Jensen LB, Hasman H. Characterization of genetic determinants of extended- spectrum cephalosporinases (ESCs) in Escherichia coli isolates from Danish and imported poultry meat. Journal Antimicrobial Chemotheraphy 2009; 64: 207-209.
- CLSI. M100-S23 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement.Wayne, PA: Clinical and Laboratory Standards Institute 2013.
- Kor SB, Choo QC, Chew CH. New integron gene arrays from multiresistant clinical isolates of members of theEnterobacteriaceaeand Pseudomonas aeruginosa from hospitals in Malaysia. Journal of Medical Microbiology 2013; 62: 412-420.
- Woolford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended spectrum β-lactamases. Journal of Antimicrobial Chemotherapy 2006; 57: 154-155.
- Cheong HT, Ho WY, Choo QC, Chew CH. β-lactamase gene blaSHV detected in bacteria isolated from retail sushi in Kampar, Malaysia. Biomedical Research 2014; 25: 25- 31.
- Rao SP, Rama PS, Gurushanthappa V, Manipura R, SrinivasanK. Extended-Spectrum β-Lactamases Producing Escherichia coli and Klebsiellapneumoniae: A Multi-Centric Study Across Karnataka. Journal of Laboratory Physicians 2014; 6(1):7-13.
- Lu SY, Zhang YL, Geng SN, Li TY, Ye ZM, Zhang DS, Zou F, Zhou HW. High diversity of extended-spectrum β- lactamase-producing bacteria in an urban river sediment habitat. Applied and Environment Microbiology 2010; 76: 5972-5976.
- Tsering DC, Das S, Adhiakari L, Pal R, Singh TS. Extended spectrum β-lactamase detection in Gram-negative bacilli of nosocomial origin. Journal of Global Infectious Diseases 2009; 1(2):87-92.
- Sahagún-Ruiz A, Granados Martinez AP, Breda LC, FragaTR, Castiblanco Valencia MM, Barbosa AS, Isaac L. Pasteurellapneumotropica evades the human complement system by acquisition of the complement regulators factor H and C4BP. PLoS One 2014; 9(10):e111194.
- Lim KT, Yasin R, Yeo CC, Puthucheary S, Thong KL. Characterization of multidrug resistant ESBL-producing Escherichia coli isolates from hospitals in Malaysia. Journalof Biomedicine and Biotechnology 2009:165637.
- Liebana E, Carattoli A,Coque TM, Hasman H, MagiorakosAP, Mevius D, Peixe L, Poirel L, Schuepbach-Regula G, Torneke K, Torren-Edo J, Torres C, Threlfall J. Public health risks of enterobacterial isolates producing extendedspectrumβ-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clinical Infectious Disease 2013; 56(7): 1030-1037.
- Parveen RM, Manivannan S, Harish BN, Parija SC. Study of CTX-M type extended spectrum β-lactamase among nosocomial isolates of Escherichia coli and Klebsiellapneumoniae in South India. Indian Journal of Microbiology 2012; 52: 35-40.
- Peerayeh SN, EslamiM, Memeriani M, Siadat SD. High prevalence of blaCTX-M-1 group extended-spectrum β-lactamase genes in Escherichia coli isolates From Tehran. Jundishapur Journal of Microbiology 2013; 6: 6863.
- Luvsansharav UO, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M, Komalamisra C, Kusolsuk T, Yamamoto Y.Prevalence of and risk factors associated with faecal carriage of CTX-M β-lactamase-producing Enterobacteriaceae in rural Thai communities. Journal of Antimicrobial Chemotherapy 2012; 67: 1769-1774.
- Quinteros M, Radice M, Gardella N, Rodriguez MM, Costa N, Korbenfeld D, Couto E, Gutkind G. Extendedspectrum β-lactamases in Enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrobial Agents and Chemotherapy 2003; 47: 2864 –2869.
- Cantón R, Coque TM. The CTX-M β-lactamase pandemic.Current Opinion in Microbiology 2006; 9: 466–475.
- Sennati S, Santella G, Di Conza J, Pallecchi L, Pino M, Ghiglione B, Rossolini GM, Radice M, Gutkind G. Changing epidemiology of extended-spectrum β-lactamases in Argentina: emergence of CTX-M-15. Antimicrobial Agents and Chemotherapy 2012; 56(11): 6003-6005.
- Jimoh SO, Shittu AA, Bello IM. Occurrence of virulence factor and extended spectrum β-lactamase in Enterobacteriaceaeassociated with ready-to-eat-fruits. International Journal of Biology and Biological Sciences 2013; 2: 83-87.
- Calbo E, Freixas N, Xercavins M, Riera M, Nicolas C, Monistrol O, SoleMdel M, Sala MR, Vila J, Garau J. Foodborne nosocomial outbreak of SHV1 and CTX-M- 15–producing Klebsiellapneumoniae: Epidemiology and control. Clinical Infectious Diseases 2011; 52: 743-749.
- Odeh R, Kelkar S, HujerAM,Bonomo RA, SchreckenbergerPC and Quinn JP. Broad resistance due to plasmidmediatedAmpC β-lactamases in clinical isolates of Escherichia coli. Clinical Infectious Diseases 2002;35(2):140-5.
- Igbinosa JH, Nwodo UU, Sosa A, Tom M, Okoh AL. Commensal Pseudomonas species isolated from wastewater and freshwater milieus in the Eastern Cape Province, South Africa, as reservoir of antibiotic resistant determinants. International Journal of Environmental Research and Public Health 2012; 9(7): 2537-2549.