ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 19
Pim-1 kinase regulates CD4+T cell differentiation
Yueming Shen1#, Yuanhong Xie2#, Yan Zhao1*, Yan Long1, Linqian Li1 and Ya Zeng1
1Department of Digestive Diseases, Changsha Central Hospital, No.163 Shaoshan Nanlu, Changsha, PR China
2Division of Gastroenterology and Hepatology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai Institute of Digestive Disease, No. 145 Shandong Zhonglu, Shanghai, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Yan Zhao
Department of Digestive Diseases
Changsha Central Hospital, PR China
Accepted on September 12, 2017
This study aimed to investigate the role of Pim-1 kinase in the regulation of naïve CD4+T cell differentiation. Naive CD4+T cells were isolated from the spleen of the mice and treated with different cytokines and Pim-1 kinase inhibitor. The expression of T-bet, GATA3, FOXP3 and RORγt proteins in naïve CD4+T cells was detected by Western blot analysis. The expression of cytokines IL-4, TGF-β, IFN- γ and IL-17 in supernatant was detected by ELISA. We found that the inhibition of Pim-1 kinase significantly reduced the expression of T-bet and ROR-γt during naïve CD4+T cells differentiation and inhibited the secretion of IFNγ and IL17 (P<0.05). In conclusion, our study suggests that Pim-1 kinase promotes the differentiation of naive CD4+T cells into Th1 and Th17 cells.
Keywords
CD4+T cell, Th1, Th17, Pim-1, Differentiation.
Introduction
The Pim-1 gene was originally identified in Murine Leukemia Virus (MuLV) induced T-cell lymphoma. The human Pim-1 gene is localized at 6P21.1-p21.31, consisted of 6 exons and 5 introns. The Pim-1 gene encodes a serine/threonine kinase, which belongs to calcium/calmodulin regulatory kinase (CAMP) [1]. Pim-1 is mainly expressed in the thymus, spleen, bone marrow and fetal liver and other hematopoietic organs, while its expression is absent in adult tissues. Recent studies have shown that Pim-1 kinase can phosphorylate a variety of protein substrates, and it is a downstream effector molecule of many cytokine signaling pathways. These cytokines can regulate the expression of Pim-1 via Signal Transducers and Activators of Transcription (STATs), which play an important role in many physiological and pathological processes such as cell proliferation, differentiation, apoptosis and tumorigenesis [2]. The structure and function of Pim-1 kinase show abnormal changes in a variety of human tumors [3-5]. The use of Pim-1 small molecule inhibitors or shRNA can inhibit tumor cell proliferation and promote apoptosis, which provides a new target for tumor therapy [6].
CD4+T cells are divided into 4 major functional subgroups of Th1, Th2, Th17 and Treg according to the differentiation and function characteristics, and the immune balance disorder among these subgroups is closely related to the occurrence and development of immune diseases such as Inflammatory Bowel Disease (IBD). Previous study reported that the levels of CD4+CD25+ Treg and Foxp3 in the peripheral blood of patients with IBD were significantly lower than those in the normal control group, and their levels were lower in IBD inflammatory activity period than in remission period [7]. In animal experiments, the decreased number or impaired function of CD4+CD25+Treg cells can lead to intestinal mucosal injury, which induced the occurrence of IBD. In addition, a large number of IL-17 secretion cells existed in the intestinal mucosa of IBD patients with acute inflammation, suggesting that Th17 cells are involved in the pathogenesis of IBD [8]. Our previous study showed that inhibiting Pim-1 in animal model of ulcerative colitis could inhibit macrophage activation and Th1, Th17-based inflammatory response [9]. Therefore, we wondered that Pim-1 may regulate the differentiation of CD4 T cells. In this study we isolated naïve CD4+T cells from the spleen of the mice and treated them with cytokines and Pim-1 inhibitor to investigate the role of Pim-1 kinase in the differentiation of naive CD4+T cells into Th1 and Th17 cells.
Materials and Methods
Isolation and culture of CD4+T cells
BALB/c mice were sacrificed and immersed in 75% ethanol for 3-5 min, and the spleen was removed on clean bench. 4 ml of mouse lymphocyte separation solution and 200 mesh nylon mesh were placed in a Petri dish. The spleen was placed on the nylon mesh and grinded with a syringe piston, and then immediately transferred to a 15 ml centrifuge tube loaded with 2 ml RPMI 1640 medium. At room temperature, T lymphocytes were separated at 2000 r/min centrifugal for 15 min, washed 2 times with PBS, and spleen CD4+T cells suspension was purified by a magnetic bead cell sorting system (Miltenyi Biotech) following the manufacturer’s protocols. The purified cells were suspended in RPMI 1640 medium supplemented with 10% fetal bovine serum, and cultured at 37°C in a humid environment containing 5% CO2 for 24 h. Trypan blue exclusion was performed to detect cell viability.
Treatment
CD4+T cells were pre-treated with 10 μmol/L or 100 μmol/L of Pim-1 kinase inhibitor (PIM-Inh) (sc-204330, Santa Cruz Corporation) for 2 h, and then mixed with CD3/CD28-beads and cultured. For the induction of Th1, cells were treated with 100 U/ml IL-2, 10 ng/ml IL-12, 10 μg/ml anti-IL4, PMA and ionomycin. For the induction of Th2, cells were treated with 10 ng/ml IL-4 and 100 U/ml IL-2. For the induction of Th17, cells were treated with 20 ng/ml IL-23, 20 ng/ml IL-6and 3 ng/ml TGF-β. For the induction of Treg, cells were treated with 5 ng/ml TGF-β and 100 U/ml IL-2. After treatment for 72 h, the cells and supernatant were collected.
Western blot analysis
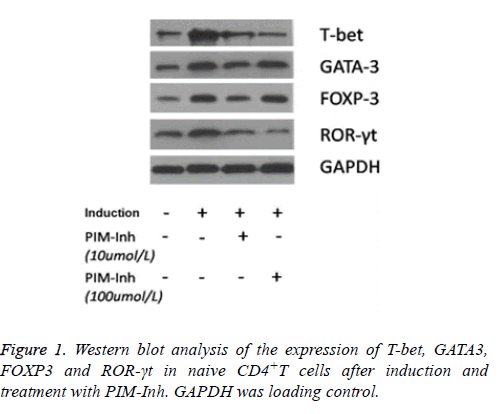
Total protein was isolated from the cells and quantitated by BSA method. 50 μg protein was loaded for 10% SDS-PAGE and transferred to PVDF membrane (Millipore, Billerica, MA, USA). Next, the membrane was incubated with primary antibodies at 4°C overnight, including ROR-γt polyclonal antibody (BioLegend Company), FOXP3 monoclonal antibody (Epitomics Company), T-bet polyclonal antibody and GATA3 polyclonal antibody (Proteintech Group). The membrane was washed with TBST for 5 min 3 times, and then incubated with secondary antibody for 1 h at room temperature. The membrane was washed with TBST for 5 min 3 times, and was developed using ECL kit (Pierce, Rockford, IL, USA) and exposed to X-ray film. Bands on X-ray films were quantified with Photoshop software.
ELISA
IFN-γ, IL-4, IL-17, TGF-β levels in cell supernatants were detected using ELISA kits (BD PharMingen) according to the manufacturer’s instructions.
Statistical analysis
The experimental data were expressed as mean ± Standard Deviation (SD), one factor analysis of variance was used to compare the data differences between groups, p<0.05 was considered statistically significant.
Results
Pim-1 inhibitor inhibited the differentiation of CD4+T cells into Th1 and Th17
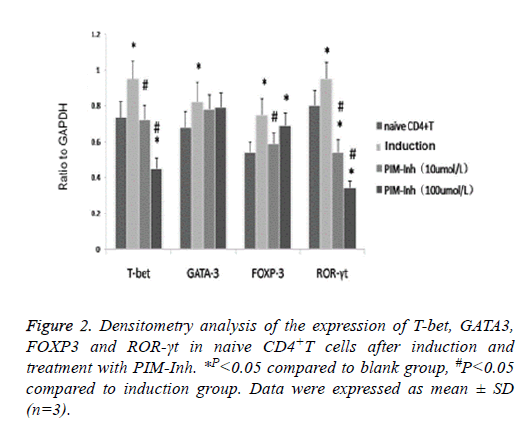
Western blot analysis showed that T-bet protein level increased significantly in CD4 T cells induced into Th1 compared to uninduced control cells, and PIM-Inh reduced the protein level of T-bet in a dose dependent manner. GATA3 protein level increased significantly in CD4 T cells induced into Th2 compared to un-induced control cells, but PIM-Inh did not significantly affect the protein level of GATA3. Similarly, FOXP3 protein level increased significantly in CD4 T cells induced into Treg compared to un-induced control cells, but PIM-Inh did not significantly affect the protein level of FOXP3. The protein level of ROR-γt increased significantly in CD4 T cells induced into Th17, and PIM-Inh inhibited the expression of ROR-γt in a dose dependent manner (Figure 1). Densitometry analysis showed that the inhibition of T-bet and ROR-γt by PIM-Inh was statistically significantly (Figure 2). Taken together, these data suggest that Pim-1 inhibitor inhibits the differentiation of CD4+T cells into Th1 and Th17.
Pim-1 inhibitor inhibited the secretion of IFNγ and IL17
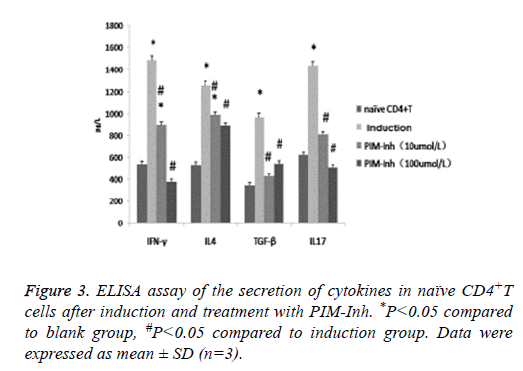
ELISA analysis showed that after induction, the secretion of IFNγ, IL4, TGF- β and IL17 all increased significantly in the differentiated CD4 T cells. However, PIM-Inh significantly inhibited the secretion of these cytokines, especially for IFNγ and IL17 (Figure 3).
Discussion
Pim-1 is mainly expressed in the thymus, spleen, bone marrow and fetal liver and other hematopoietic organs [1,2]. In our previous study we found that Pim-1 was related to Th1 and Th17 based inflammatory response during ulcerative colitis [9]. In this study, we isolated naïve CD4+T cells from the spleen of the mouse in which Pim-1 is expressed, and then used different doses of PIM-Inh to inhibit Pim-1 for pretreatment, 2 h later different agents were used to induce the differentiation of CD4+T cells. Compared with the control group, the corresponding nuclear transcription factor and cytokines increased significantly in the induced group, suggesting that the differentiation of naïve CD4+T cells was successfully induced. After treatment with PIM-Inh, T-bet, ROR-γt, IFNγ and IL17 levels were significantly inhibited, suggesting that blocking Pim-1 could inhibit the differentiation of naive CD4+T to Th1 and Th17, but had no obviously effect on the expression of FOXP3 and GATA-3. These results suggest that Pim-1 is involved in the differentiation of Th1 and Th17.
Recently, it was reported that the use of highly selective Pim1/3 small molecule inhibitor could block the proliferation of lymphocytes in G0/G1 phase, but did not induce T cell apoptosis [10]. Shin et al. found that Pim-1 was activated in asthma and allergic reaction, and Pim-1 inhibitor could significantly reduce airway hyperresponsiveness and airway inflammation [11]. Runx3 plays an important role in regulating Th1/Th2 cells balance, and the regulation of Th cell differentiation by Pim-1 may involve RUNX family proteins because knockout of Runx3 gene in the mice could cause spontaneous inflammation of the intestine [12]. In this study, intestinal cytokines were detected and we found that the expression of Th1 type cytokine IFN-γ increased significantly, Th2 type cytokine IL-4 only increased slightly, while cytokines IL-10 and TGF-β did not change obviously. In addition, we found that the expression of Th1 transcription factor T-bet and Th2 transcription factor GATA-3 increased significantly, suggesting that Runx3 could regulate the differentiation of Th cells by regulating the expression of T-bet and GATA-3. Kitoh et al. found that RUNX1 was an important transcription factor that maintained Treg cell function, and NFκB and NFAT were transcription factors for the expression of Th1 transcription factor T-bet and cytokine IFNγ [13-15].
Taken together, our study suggests that Pim-1 kinase promotes the differentiation of naïve CD4+T cells into Th1 and Th17 cells, and blocking Pim-1 may have important implication for the treatment of inflammatory response.
Acknowledgements
This work did not receive any funding or support.
Disclosure of Conflict of Interest
None
References
- Padma R, Nagarajan L. The human PIM-1 gene product is a protein serine kinase. Cancer Res 1991; 51: 2486-2489.
- Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol 2005; 37: 726-730.
- Abdel-Magid AF. Pim kinase inhibitors for the treatment of cancer and possibly more. ACS Med Chem Lett 2014; 5: 730-731.
- Santio NM, Eerola SK, Paatero I, Yli-Kauhaluoma J, Anizon F, Moreau P, Tuomela J, Härkönen P, Koskinen PJ. Pim kinases promote migration and metastatic growth of prostate cancer xenografts. PLoS One 2015; 10: 130340.
- Zhu X, Xu JJ, Hu SS, Feng JG, Jiang LH, Hou XX, Cao J, Han J, Ling ZQ, Ge MH. Pim-1 acts as an oncogene in human salivary gland adenoid cystic carcinoma. J Exp Clin Cancer Res 2014; 33: 114.
- Shah N, Pang B, Yeoh KG, Thorn S, Chen CS, Lilly MB, Salto-Tellez M. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer 2008; 44: 2144-2151.
- MacDonald TT, Vossenkaemper A, Fantini M, Monteleone G. Reprogramming the immune system in IBD. Dig Dis 2012; 30: 392-395.
- Boden EK, Snapper SB. Regulatory T cells in inflammatory bowel disease. Curr Opin Gastroenterol 2008; 24: 733-741.
- Liu ZJ, Yadav PK, Su JL, Wang JS, Fei K. Potential role of Th17 cells in the pathogenesis of inflammatory bowel disease. World J Gastroenterol 2009; 15: 5784-5788.
- Shen YM, Zhao Y, Zeng Y, Yan L, Chen BL, Leng AM, Mu YB, Zhang GY. Inhibition of Pim-1 kinase ameliorates dextran sodium sulfate-induced colitis in mice. Dig Dis Sci 2012; 57: 1822-1831.
- Jackson LJ, Pheneger JA, Pheneger TJ, Davis G, Wright AD, Robinson JE, Allen S, Munson MC, Carter LL. The role of PIM kinases in human and mouse CD4+ T cell activation and inflammatory bowel disease. Cell Immunol 2012; 272: 200-213.
- Shin YS, Takeda K, Shiraishi Y, Jia Y, Wang M, Jackson L, Wright AD, Carter L, Robinson J, Hicken E, Gelfand EW. Inhibition of Pim1 kinase activation attenuates allergen-induced airway hyperresponsiveness and inflammation. Am J Respir Cell Mol Biol 2012; 46: 488-497.
- Wang M, Okamoto M, Domenico J, Han J, Ashino S, Shin YS, Gelfand EW. Inhibition of Pim1 kinase prevents peanut allergy by enhancing Runx3 expression and suppressing T(H)2 and T(H)17 T-cell differentiation. J Allergy Clin Immunol 2012; 130: 932-944.
- Qiao J, Liu Y, Wu Y, Li X, Zhu F, Xia Y, Yao H, Chu P, Li H, Ma P, Li D, Li Z, Xu K, Zeng L. Aberrant expression of RUNX3 in patients with immune thrombocytopenia. Int Immunopharmacol 2015; 28: 252-256.
- Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 2009; 31: 609-620