ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 10
Plasminogen: a potential target gene for dietary supplements and biomarker of the early stage of obesity by fatigue mice
Li Zheng*
Department of Pharmacy, Xingtai Medical College, Xingtai, PR China
- *Corresponding Author:
- Li Zheng
Department of Pharmacy
Xingtai Medical College, PR China
E-mail: lizheng_come_34@163.com
Accepted date: February 02, 2017
Background: The Early Stage of Obesity by Fatigue (ESOF) is a serious sub-health issue due to the social burden and a leading cause of other diseases even death. Accordingly, a broad range of dietary supplements have been researched and applied in an effort to treat ESOF. However, only a small number of dietary supplements have been specifically applied for gene diagnosis and the targeted gene therapy for early treatment of obesity by fatigue.
Materials and Methods: In order to search the target genes for the examination in this research, GSE 038921 was downloaded from the public database, Gene Expression Omnibus, including all three samples of test and control groups. Analysis was carried out using R software packages, protein-protein interaction network, Kyoto Encyclopaedia of Genes and Genomes pathway and Gene ontology enrichment. It yielded a total of six key genes caused by limited sleep. The result was tested and verified by other data: GSE 65557 and GSE 34959.
Results: Finally, one gene and one key pathway were identified: Plg; complement and coagulation cascades.
Conclusions: This study suggested target gene and biological marker for treatments of ESOF by dietary supplements, providing a new perspective on the pathological process of the early phrase of ESOF.
Keywords
Obesity, Complement and coagulation cascades, Plasminogen, Dietary supplements, Biomarker
Introduction
The early stage of obesity by fatigue (ESOF) is a former stage of obesity caused by sleep deprivation and over-fatigue [1]. It is responsible for numerous global public health issues. It not only leads to obesity and the social and economic burden, but can also potentially culminate in an increased risk of other serious diseases; furthermore, karoshi may be the most serious outcome of ESOF [1,2]. Meanwhile, obese individuals gain poor sleep and more significant fatigue than others [3]. Hence, the relevant preventive treatments are an urgent matter.
The dietary supplements used to treat ESOF can be categorised into four groups: traditional Chinese medicine, polyphenols extracted from natural flora and fauna, food and others. Firstly, Chinese ginseng and protopanaxatriol which exemplify the traditional Chinese medicine [4,5] for reducing obesity in high fat diet-induced mice. Secondly, polyphenols plays important roles in lessen obesity through accelerating fat metabolism, such as flavonoids (anthocyanins and flavanols, isoflavones, dihydroflavonoids, flavonoids), organic acids, tannin [6]. Long-term purified betacyanins from Hylocereus undatus peel and cocoa flavanol oligomers in taking reduced body weight gain in HFD-induced mice [7,8], short-term anthocyanin-rich bilberry extract without the flavanol epicatechin plays the same role [9]. Soy isoflavones ameliorates the accumulation of hepatic lipid droplet formation in rats [10]. The Fortunella margarita fruit extract effects similarly [11]. The flavonoids of isorhamnetin decreased hepatic steatosis and obesity in HFDinduced mice [12]. Aged citrus peel (chenpi) extract (5- demethylated polymethoxyflavones) as a kind of andorganic acids has a certain effect on prevention of obesity [13]. The tannin including gallates and green tea catechins also has the benefit of prevention obesity [6]. Thirdly, the mice gain lower weight by fed with food including oat or wheat bran fiber than without including dietary fiber, furthermore the rice bran enzymatic extract plays role in protecting the microvascular in obesity. Additionally, the ob/ob mice also gain benefit from Dallulosea in losing body and liver weights [14-16]. Fourthly, there are some other dietary supplements, such as 4-hydroxy-2, 2, 6, 6-tetramethylpiperidine-N-oxyl radical, (a nitroxide compound) [17], Yacon (Smallanthus sonchifolius) [18], glutamine [19], Omega-3 phospholipids from fish [20], Betaine [21] prevent the accumulation of whole-body and hepatic lipid in High Fat Diet (HFD) induced mice. All of these have effect on obesity, however, the effects were not fully stable [6], the mechanism need to be explored for searching more dietary supplements.
In order to overcome this problem, some scientists have sought to address this condition from a molecular biology standpoint. Some studies have claimed that Peroxisome Proliferator-Activated Receptors (PPARs) are important pathways. More attention is paid by people to peroxisome proliferator-activated receptor γ (PPARγ) or PPARγ activator 1-α for improving obesity by dietary supplements, for PPARγ is an important gene in adipocyte differentiation and maturation. Furthermore, the pathway of PPARs is also important in pathology of obesity. Additional, there are some other key genes in preventing obesity by dietary supplements, such as peroxisome proliferator-activated receptor α (PPARα) [11,14], PPARγ [10,12,14], activated protein kinase in adipose tissue [13], CCAAT-Enhancer-Binding Protein α (C/EBPα) [4,6], Sterol Regulatory Element-Binding Protein 1 (SREBP-1) [6,14], FSP27 [10], FGF21-related genes [8,21], NO [15], leptin and adiponectin [17], short chain fatty acids [18], liver X receptor alpha (LXRα), ATP-binding cassette transporter (ABCA1), adenosine monophosphate-activated protein kinase (AMPactivated protein kinase), and other regulate adipogenesis [6]. Nonetheless, these researches initiatives have certain limitations because of being short of exploring other processes of pathology in ESOF excluding fat metabolism disorders [6]. More accurate target genes are needed to satisfy the demands for further research.
In short, these studies focus on fat metabolism. However, the pathological processes of ESOF may include other unknown changes. The research on the genes of ESOF was lack. Relevant research conditions of obesity generally are high-fat food and cold, yet the research on sleep limit is largely inadequate. However, as has been asserted by Yarandi et al. sleep deprivation can lead to obesity [1]. Additionally, whilst obesity ultimately leads to chronic diseases, ESOF is still somewhat unknown. Accordingly, this research seeks to address this gap in the existing research.
This study attempted to determine the key genes leading to ESOF of mice based on a certain degree of sleep deprivation. Moreover, the pathological change of epididymal WAT of mice of former state also is examined. Therefore, the expressive data of GSE 038921 was downloaded, whilst the different genes (DEGs) were analysed by Limma package in R. Subsequently, the key genes can be determined by constructing the protein network and functional pathways with the help of DEGs and verification with the help of other data. In conclusion, this study provided potential targets for medicine and the potential combination of biomarkers, thereby enhancing understanding of the pathological process of ESOF.
Materials and Methods
Materials (Data source)
Genes expressions data of GSE 038921 (financed by the German Research Foundation (DFG; SFB 654) and the Max Planck Society) [22] was downloaded from GEO based on the GPL8321 platform: [Mouse430A_2] Affymetrix Mouse Genome 430A 2.0 Array. GSE 038921 includes three control group samples and three Timed Sleep Restriction (TSR) samples whose daily sleep time were restricted to six hours by gently dealing with for five uninterrupted days and that were killed at Zeitgeber time 18 on the final day. There was no significant difference in body weight between the groups in experiment, but there was a trend of obesity during the recovery period [22], we believed that this period belonged to the ESOF.
DEGs analysis
The original matrix data was corrected and normalised. Comparative analysis of DEGs was carried out through the logarithm expression of corrected data by the Limma [23] package in R. The p value<0.05 (corrected by FDR (false discovery rate)) and values of |log2-fold change (log2-FC)|>1 were considered as the cut-off criteria. Subsequently, the volcano plot was constructed using DEGs.
Networks construction
The Protein-Protein Interaction (PPI) network which formed the DEGs was developed using the online tool, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://www.string-db.org) [24]. The hob nodes were extracted from the downloaded string interactions by R. Thereafter, related data was downloaded to construct the sub-network by using the MCODE [25] plugin for Cytoscape: the default parameters: degree cut-off ≥ 2, node score cut-off ≥ 0.2, K-core ≥ 2, and maximum depth=100. Finally, the sub-network containing the hob nodes was selected as the object of the next study.
Pathway analysis
In order to investigate the changes of epididymal WAT in ESOF from the perspective of functional level, pathway enrichment analyses were performed. This included GO biological function (GO BF) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis. GO BF enrichment analyses were performed for the DEGs in the sub-network using the online tool, DAVID (the database for annotation, visualisation, and integrated discovery, https:// david.ncifcrf.dov) [26]. The p value<0.05 (corrected by FDR) was set as the cut-off condition, and was visualised by ggplot2 [27] package in R. Meanwhile, the DEGs were enriched in KEGG pathway and analysed by cluster Profiler [28] package in R. The analysis by p value in KEGG was the same with the analysis in GO BF. Furthermore, the common genes in GOterm enrichment, KEGG pathway enrichment and the hub nodes in the PPI network were selected for the following analysis.
Verification
The selected genes were verificated by GSE 39549 (Dietinduced obesity model: white adipose tissue, 40 sample) and GSE 65557 (Short-term high fat diet effect on epididymal adipose fractions: time course, 18 sample) by data analysis tools of GEO online so as to test and verify the selected genes more accurately. The selected times were 0 day, 14 days in GSE395490 day; 0 day, 7 days in GSE 65557, GSE 34959. Comparative analysis of DEGs was performed through the logarithm expression of related on-line data by T-test using trial version of SPSS19.0. (p value<0.05 was the comparative criteria).
Results
Identification of DEGs
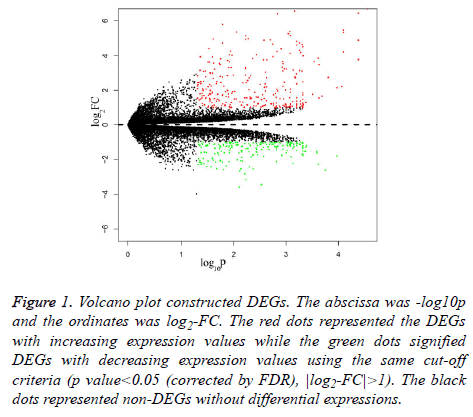
For dataset GSE 038921, 483 DEGs were identified in total at | log2-FC|>1 and p value<0.05 (corrected by FDR) between the three control and three TSR group samples. Besides these, there were 278 increasedly regulatory DEGs and 205 decreasedly regulatory DEGs. The varied DEGs were visualised by the volcano plot (Figure 1).
Figure 1: Volcano plot constructed DEGs. The abscissa was -log10p and the ordinates was log2-FC. The red dots represented the DEGs with increasing expression values while the green dots signified DEGs with decreasing expression values using the same cut-off criteria (p value<0.05 (corrected by FDR), |log2-FC|>1). The black dots represented non-DEGs without differential expressions.
Networks’ construction of DEGs
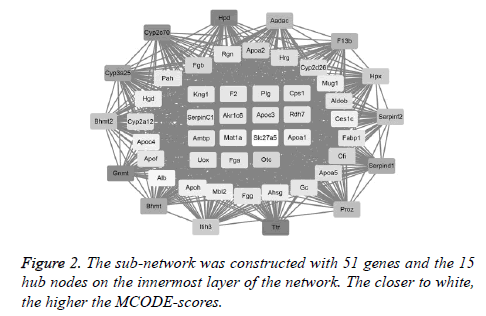
The first network (PPI network) containing 483 DEGs was constructed by STRING software to excavate the interactive products. Notably, the top 15 hub nodes, such as Kng1 and F2, extracted from the downloaded string_interactions table, are shown in Figure 2. In total, 15 separate interaction networks were identified. As only the first sub-network was constructed with 51 DEGs, including the 15 hub nodes, it was selected as the subject for the next bioinformatics analysis (Figure 2).
Pathway analyses of DEGs
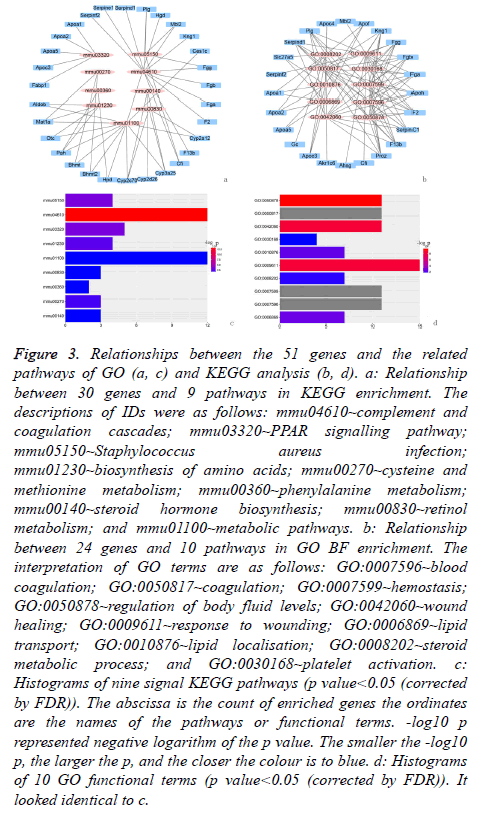
GO BF analysis of the 51 DEGs in the selected biological process indicated that there were 10 varying biological progresses. The result visualised by forth-putting ggplot2 (package in R) showed that coagulation and the fat metabolic process were the key biological processes (Figures 3a and 3b).
Figure 3: Relationships between the 51 genes and the related pathways of GO (a, c) and KEGG analysis (b, d). a: Relationship between 30 genes and 9 pathways in KEGG enrichment. The descriptions of IDs were as follows: mmu04610~complement and coagulation cascades; mmu03320~PPAR signalling pathway; mmu05150~Staphylococcus aureus infection; mmu01230~biosynthesis of amino acids; mmu00270~cysteine and methionine metabolism; mmu00360~phenylalanine metabolism; mmu00140~steroid hormone biosynthesis; mmu00830~retinol metabolism; and mmu01100~metabolic pathways. b: Relationship between 24 genes and 10 pathways in GO BF enrichment. The interpretation of GO terms are as follows: GO:0007596~blood coagulation; GO:0050817~coagulation; GO:0007599~hemostasis; GO:0050878~regulation of body fluid levels; GO:0042060~wound healing; GO:0009611~response to wounding; GO:0006869~lipid transport; GO:0010876~lipid localisation; GO:0008202~steroid metabolic process; and GO:0030168~platelet activation. c: Histograms of nine signal KEGG pathways (p value<0.05 (corrected by FDR)). The abscissa is the count of enriched genes the ordinates are the names of the pathways or functional terms. -log10 p represented negative logarithm of the p value. The smaller the -log10 p, the larger the p, and the closer the colour is to blue. d: Histograms of 10 GO functional terms (p value<0.05 (corrected by FDR)). It looked identical to c.
The result of KEGG analysis, in an effort to obtain a more comprehensive understanding of the pathways of 51 DEGs, was nine enriched pathways, which were showed in Figures 3c and 3d.
The quantity of the hot DEGs by the analysis of KEGG pathway, GO enrichment and PPI network was shown in Table 1.
| KEGG | GO | PPI network | |
|---|---|---|---|
| Quantity of the DEGs | 30 | 24 | 15 |
Table 1. Quantity of the hot DEGs by the analysis of KEGG pathway, GO enrichment and PPI network.
Six genes were mainly enriched in two key pathway complement and coagulation cascades, PPAR signalling pathway): Apoa1, Apoc3, Kng1, F2, Fga and Plg. As such, these six genes are located where the three circles, including hub nodes, GO terms and KEGG pathways, overlap in the Venn diagram (Table 2).
| Complement and coagulation cascades | PPAR signalling pathway | |||||
|---|---|---|---|---|---|---|
| Genes | F2 | Kng1 | Fga | Plg | Apoc3 | Apoa1 |
Table 2. The six key genes and two key pathways in epididymal WAT of ESOF.
Verification
One different gene (Plasminogen: Plg) enriched in complement and coagulation cascades was involved in both two data (Table 3).
| Apoa1 | Apoc3 | F2 | Kng1 | Fga | Plg | |
|---|---|---|---|---|---|---|
| GSE 65557 | 0.372↓ (n=3) | 0.167↓ (n=3) | 0.979↑ (n=3) | 0.707↓ (n=3) | 0.933↓ (n=6) | 0.045↑* (n=3) |
| GSE 39549 | 0.033↑* (n=4) | 0.478↑ (n=4) | 0.654↑ (n=4) | 0.267↑ (n=12) | 0.035↑* (n=8) | 0.041↑* (n=16) |
Table 3. Verifying the different genes in data GSE 65557 and GSE 39549.
Discussion
This research discovered that one key gene in ESOF in mice: Plg, which was gathered in one important way: complement and coagulation cascades pathway. Plg, normally decreases in obesity, but increase in ESOF in mice in our research. This view was first put forward as we known. We hold that obesity can lead to reduced Plg, which cause thrombosis, but in ESOF Plg is activated with a state of body excitement.
Plg, with another name: plasminogen has coagulation and noncoagulation effects. Plg is a key gene in the process of complement and coagulation cascades, for it is the precursor of plasmin. Whilst, Plg has close relationgships of obesity and inflammatory. For example the gonadal fat depots in the Plg-/- mice was higher compared with the wild-type mice by fed with the same high-fat diet [29], that is to say Plg may has negative effect on obesity induced by high-fat diet. Similarly, Al Salmi et al. reported that Plg was negatively related with human birth weight among thousands participants [30].
We hypothesized that angiogenesis may be one of the potential pathology of ESOF. Plg also increased on the conditions of sleep-lacked, fat-diet [31], and cold [32]. In the last two of these the white adipose tissue was activated, which result in generation of vessel accompanied with heat. Plg has positive effect on the process of angiogenesis in the skin wound-healing model [33]. Additionally, plasmin, the activated product of Plg, may induce inflammation by activation related cells [34]. However, Cho et al. reported that a fragment of human Plg, had negative effect on angiogenesis [35]. We take the time order and the complexity of coagulation system as the reason. We believe that the pathological process in ESOF of mice is unique, although chronic inflammation takes place in the latter stages of obesity.
The unique pathological process of ESOF is most likely caused by beige fat (with brown fat function and white fat appearance) contained in WAT of the experimental material. Since epididymal tissue is the general organ storing beige fat, and there is no connective tissue between the two, individuals can easily acquire beige fat when sampling WAT [32,36]. Meanwhile, beige fat can reconstruct vessels, generate and burn fat, which is consistent with the pathological process. Ultimately, obesity is the final fate of mice regardless of the paradoxical fat metabolism. The researcher speculates that the brown and activation of beige fat is one of the features of early pathological process of mice ESOF.
The research of this study can assist the clinic diagnosis and prevent ESOF by suitable food supplements. It can be used as biomarker and one filter to select effective food by the relationship of the target gene. Meanwhile, it is suitable for crowds who have various fat complications and cardiovascular diseases with the same pathological basis.
Nonetheless, the gene still needs more evidences to support the result.
Conclusion
In summary, this study discovered that the pathology in epididymal WAT of ESOF was primarily the result of vascularrelated changes. Gene enriched in complement and coagulation cascades were defined, Plg. Ultimately, this research may facilitate a deeper understanding of the pathological process of ESOF as well as highlight a potential biomarker and target for ancillary diagnosis and treatment. However, the samples size was small, thus further investigations are required to substantiate the conclusions reached.
Acknowledgements
None.
Conflict of Interest
The author declares no conflicts of interest affected the credibility or reliability of this research.
References
- Logue EE, Scott ED, Palmieri PA, Dudley P. Sleep duration, quality, or stability and obesity in an urban family medicine center. J Clin Sleep Med 2014; 10: 177-182.
- Li J. Karoshi: An international work-related hazard? Int J Cardiol 2016; 206: 139-140.
- Jarosz PA, Davis JE, Yarandi HN, Farkas R, Feingold E, Shippings SH, Smith AL, Williams D. Obesity in urban women: associations with sleep and sleepiness, fatigue and activity. Womens Health Issues 2014; 24: e447-e454.
- Li X, Luo J, Anandh BPV, Zhang W, Gilbert E, Cline M, McMillan R, Hulver M, Alkhalidy H, Zhen W, Zhang H, Liu D. Dietary supplementation of Chinese ginseng prevents obesity and metabolic syndrome in high-fat diet-fed mice. J Med Food 2014; 17: 1287-1297.
- Zhang Y, Yu L, Cai W, Fan S, Feng L. Protopanaxatriol, a novel PPAR- γ antagonist from Panax ginseng, alleviates steatosis in mice. Sci Rep 2014; 4: 7375.
- Wang S, Moustaid-Moussa N, Chen L, Mo H, Shastri A. Novel insights of dietary polyphenols and obesity. J Nutr Biochem 2014; 25: 1-18.
- Dorenkott MR, Griffin LE, Goodrich KM, Thompson-Witrick KA, Fundaro G, Ye L, Stevens JR, Ali M, OKeefe SF, Hulver MW, Neilson AP. Oligomeric Cocoa procyanidins possess enhanced bioactivity compared to monomeric and polymeric Cocoa procyanidins for preventing the development of obesity, insulin resistance, and impaired glucose tolerance during high-fat feeding. J Agric Food Chem 2014; 62: 2216-2227.
- Song H, Chu Q, Xu D, Xu Y, Zheng X. Purified Betacyanins from Hylocereus undatus peel ameliorate obesity and insulin resistance in high-fat-diet-fed mice. J Agric Food Chem 2016; 64: 236-244.
- van der Heijden RA, Morrison MC, Sheedfar F, Mulder P, Schreurs M, Hommelberg PP, Hofker MH, Schalkwijk C, Kleemann R, Tietge UJ, Koonen DP, Heeringa P. Effects of anthocyanin and flavanol compounds on lipid metabolism and adipose tissue associated systemic inflammation in diet-induced obesity. Mediators Inflamm 2016; 2016: 2042107.
- Xiao CW, Wood CM, Weber D, Aziz SA, Mehta R, Griffin P, Cockell KA. Dietary supplementation with soy isoflavones or replacement with soy proteins prevents hepatic lipid droplet accumulation and alters expression of genes involved in lipid metabolism in rats. Genes Nutr 2014; 9: 1-12.
- Tan S, Li M, Ding X, Fan S, Guo L. Effects of Fortunella margarita fruit extract on metabolic disorders in high-fat diet-induced obese C57BL/6 mice. PLoS One 2014; 9: e93510.
- Zhang Y, Gu M, Cai W, Yu L, Feng L. Dietary component isorhamnetin is a PPARγ antagonist and ameliorates metabolic disorders induced by diet or leptin deficiency. Sci Rep 2016; 6: 19288.
- Guo J, Tao H, Cao Y, Ho CT, Jin S. Prevention of obesity and type 2 diabetes with aged citrus peel (Chenpi) extract. J Agric Food Chem 2016; 64: 2053-2061.
- Han S, Jiao J, Zhang W, Xu J, Wan Z, Zhang W, Gao X, Qin L. Dietary fiber prevents obesity-related liver lipotoxicity by modulating sterol-regulatory element binding protein pathway in C57BL/6J mice fed a high-fat/cholesterol diet. Sci Rep 2015; 5: 15256.
- Justo ML, Claro C, Vila E, Herrera MD, Rodriguez-Rodriguez R. Microvascular disorders in obese Zucker rats are restored by a rice bran diet. Nutr Metab Cardiovasc Dis 2014; 24: 524-531.
- Itoh K, Mizuno S, Hama S, Oshima W, Kawamata M, Hossain A, Ishihara Y, Tokuda M. Beneficial effects of supplementation of the rare sugar d-allulose against hepatic steatosis and severe obesity in lepob/lepob mice. J Food Sci 2015; 80: 1619-1626.
- Kim CHJ, Mitchell JB, Bursill CA, Sowers AL, Thetford A, Cook JA, van Reyk DM, Davies MJ. The nitroxide radical TEMPOL prevents obesity, hyperlipidaemia, elevation of inflammatory cytokines, and modulates atherosclerotic plaque composition in apoE -/-, mice. Atherosclerosis 2015; 240: 234-241.
- Caetano BF, Moura NA, Almeida AP, Dias MC, Sivieri K, Barbisan LF. Yacon (Smallanthus sonchifolius) as a food supplement: health-promoting benefits of fructooligosaccharides. Nutrients 2016; 8: 436.
- Laviano A, Molfino A, Lacaria MT, Canelli A, De Leo S, Preziosa I, Rossi Fanelli F. Glutamine supplementation favors weight loss in non-dieting obese female patients. A pilot study. Eur J Clin Nutr 2014; 68: 1264-1266.
- Rossmeisl M, Medrikova D, Van Schothorst EMV, Pavlisova J, Kuda O, Hensler M, Ova KB, Flachs P, Stankova B, Vecka M, Tvrzicka E, Zak A, Keijer J, Kopecky J. Omega-3 phospholipids from fish suppress hepatic steatosis by integrated inhibition of biosynthetic pathways in dietary obese mice. Biochimica et Biophysica Acta 2014; 1841: 267-278.
- Ejaz A , Martinez-Guino L, Goldfine AB, Ribas-Aulinas F, De Nigris V, Ribo S, Gonzalez-Franquesa A, Garcia-Roves PM, Li E, Dreyfuss JM, Gall W, Kim JK, Bottiglieri T, Villarroya F, Gerszten RE, Patti ME, Lerin C. Dietary betaine supplementation increases fgf21 levels to improve glucose homeostasis and reduce hepatic lipid accumulation in mice. Diabetes 2016; 65: 902-912.
- Husse J, Hintze S C, Eichele G, Lehnert H, Oster H. Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. PLoS One 2012; 7: e52983.
- Yu G, Wang LG, Han Y, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284-287.
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins globally integrated and scored. Nucleic Acids Res 2011, 39: 561-568.
- Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY, Arai AE. Multi Contrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012; 14: 83.
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44-57.
- Wickham H. ggplot2: elegant graphics for data analysis. Switzerland: Springer Publishing Ltd (2nd edn.) 2016.
- Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013; 29: 1830-1831.
- Hoover-Plow J, Ellis J, Yuen L. In vivo plasminogen deficiency reduces fat accumulation. Thromb Haemost 2002; 87: 1011-1019.
- Al Salmi, Hoy WE, Kondalsamy-Chennakesavan S, Wang Z, Gobe GC, Barr EL, Shaw JE. Disorders of glucose regulation in adults and birth weight: results from the Australian Diabetes, Obesity and Lifestyle (AUSDIAB) Study. Diabetes Care 2008; 31: 159-164.
- Voigt A, Agnew K, van Schothorst EM, Keijer J, Klaus S. Short-term, high fat feeding-induced changes in white adipose tissue gene expression is highly predictive for long-term changes. Mol Nutr Food Res 2013; 57: 1423-1434.
- Xue Y, Petrovic N, Cao R, Larsson O, Lim S. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metab 2009; 9: 99-109.
- Chen PK, Chang BI, Kuo CH, Chen PS, Cho CF. Thrombomodulin functions as a plasminogen receptor to modulate angiogenesis. FASEB J 2013; 27: 4520-4531.
- Syrovets T, Lunov O, Simmet T. Plasmin as a proinflammatory cell activator. J Leukoc Biol 2012; 92: 509-519.
- Cho CF, Chen PK, Chang PC, Wu HL, Shi GY. Human plasminogen kringle 1-5 inhibits angiogenesis and induces thrombomodulin degradation in a protein kinase A-dependent manner. J Mol Cell Cardiol 2013; 63: 79-88.
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013; 19: 1252-1263.