ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 15
Polybutylcyanoacrylate nanoparticles loaded with antisense oligodeoxynucleotide of hTERT inhibit glioma growth
1Department of Neurosurgery, the Second Affiliated Hospital of Soochow University, Suzhou 215004, PR China
2Changyi People’s Hospital, Changyi, China, Changyi 261300, PR China
#These authors contributed equally to this work
Accepted date: July 2, 2017
Objective: To prepare hTERT antisense oligodeoxynucleotide (ASODN) loaded polybutylcyanoacrylate nanoparticles (PBCA-NP) and evaluate the efficacy to inhibit glioma growth in vitro.
Methods: hTERT ASODN-PBCA-NP was prepared using the emulsion polymerization method. The size and morphology were observed by transmission electron microscopy. The drug loading and entrapment efficiency of ASODN in the nanoparticles were measured by UV spectra. The viability of SHG44 glioma cells was detected by MTT assay, cell cycle was determined by flow cytometry. The expression of hTERT mRNA was measured by RT-PCR, telomerase protein expression was measured by immunocytochemical analysis.
Results: The nanoparticles were discrete and uniform spheres with average diameter of 120 nm, with drug loading and entrapment efficiency of 71.17% and 95.20%, respectively. Zeta potential was +41.3 mV. SHG44 cells treated by ASODN-NP showed reduced hTERT mRNA and reduced telomerase protein expression levels, reduced cell viability, increased percentages of cells at G0/G1 and G2/M phases and decreased percentages of cells at S phase compared to control groups.
Conclusion: PBCA nanoparticles loaded with antisense oligodeoxynucleotide of hTERT can inhibit the expression of hTERT and inhibit the viability of glioma cells.
Keywords
hTERT, Polybutylcyanoacrylate, Nanoparticles, Antisense oligodeoxynucletide, Emulsion polymerization.
Introduction
Glioma is the most common aggressive primary malignant brain tumors. Although significant advances have been made by neurosurgical operative techniques, adjuvant chemotherapy and radiotherapy, the prognosis of glioma remains unfavourable and median survival is within 2 years. The bloodbrain barrier (BBB) consists of endothelial cells which are connected by tight junction, and many enzymes, receptors, transporters and efflux pumps of multidrug resistance pathways. As a result, most chemotherapy agents are unable to reach the glioma in therapeutic concentrations [1]. Nanoparticles made by poly (butyl cyanoacrylate) (PBCA) or poly (lactic-co-glycolicacid) (PLGA) with surface modified by polysorbate 80 enable the transport of chemotherapeutic agents such as doxorubicin across BBB and enter the tumor [2]. After intravenous injection of nanoparticles loading doxorubicin to intracranial glioma bearing rats, the life span of the animals significantly increased. Moreover, these particles considerably reduced the Dox induced cardiotoxicity and testicular toxicity.
It has been found that more than 50% gliomas have increased telomerase activity, and the expression of telomerase is correlated with the grade of malignancy [3]. The level of hTERT mRNA is predictive of glioma patient survival. Antisense oligodeoxynucleotide (ASODN) could inhibit or block the expression of targeting gene, resulting to the loss of activity. It is difficult for ASODN to penetrate biomembrane as a hydrophilic anionic polymer, which can be biodegraded by enzymes in vivo, and its low biostability limited the applications. PBCA nanoparticles are considered to have relatively low toxicity, moreover, it is among the most rapidly biodegrading synthetic polymers [4]. In this study, we prepared hTERT ASDO loaded PBCA nanoparticles (NP) by emulsion polymerization method and evaluated the efficacy to inhibit glioma in vitro.
Materials and Methods
Preparation of PBCA-NP and SODN-NP/ASODN-NP
The PBCA-NP was prepared using the emulsion polymerization method as described previously [5]. Briefly, 0.12 g DEAE-Dextran and 0.18 g Dextran70 were dissolved in 25 ml hydrochloric acid solution, and pH is 2.0. 0.19 ml BCA monomer was slowly dropped into the mixed solution and continued stirring for 6 h, and then filtered by a sintered glass funnel with diameter 0.45 μm. 1 mol/L NaOH was added into such solution until pH reached 7.0 to get PBCA-NP. A certain amount of sense or antisense oligonucleotides was added into the above nanoparticle fluid and continue to stir for 3 h to get SODN-NP/ASODN-NP. hTERT ASODN was designed according to the sequences of hTERT mRNA (AF015950), the sequences were as follows: ASODN1 5’- GCA CGG CTC GGC AGC GGGA-3’; ASODN2 5’- GGAGCG CGC GGC ATC GCGGG-3’; ASODN3 5’-GGT AGA GAC GTG GCT CTGA-3; Sequence of 5’ end FITC labeled antisense oligonucleotides (FASODN) was same as ASODN1; Antisense oligonucleotides sequence (SODN) was designed as control: 5’- CCC GCG ATG CCGCGC GCT CC-3’.
Characterization of NP and SODN-NP/ASODN-N-NP
The NP or SODN-NP/ASODN-N-NP was diluted with distilled water and measured by Laser diffraction particle size analyzer (Mastersizer 3000HS) to assess size and size distribution of nanoparticles. The NP or Dox-NP Suspension was diluted appropriately and dropped on the carbon film coated copper net, stained by 2% phosphate tungsten, dried naturally for 30 min, then observed by Transmission electron microscope (TEM, JEM-100CX, Japan) for its shape and size. The freeze dried SODN-NP/ASODN-N-NP powder which was kept for three months under normal temperature was dissolved by distilled water to detect its stability.
The Drug loading and entrapment efficiency of ODN in the nanoparticles were measured by means of UV spectra at weave length of 260 nm. ODN standard solution was prepared with the concentration gradient of 0.25, 0.5, 0.75, 1.0, 1.25, 2.5, 5.0 and 10 μg/ml. Then 5 ml ODN-PBCA-NP emulsion was centrifuged by speeding cold Freeze (LABCONCO Free Zone 12 L freeze dryer, USA) at 20,000 r/min for 30 min, then the supernatant was taken and the absorbance was measured at the wavelength of 481 nm by UV Spectrophotometry. The drug loading ratio (DL) and encapsulation efficiency (EE) were calculated as follows [6]: DL%=ODNtotal-ODNsupernatant/BCA × 100%; EE=ODNtotal-ODNsupernatant/ODNtotal × 100%.
NP-ODN was suspended in a dialysis system and the released ODN was detected by UV Spectrophotometry.
In vitro cytotoxicity assay
SHG44 glioma cells were seeded into 96-well culture plates at a density of 5 × 104 cells/well and maintained in 100 μL of DMEM medium supplemented with 10% FBS at 37°C in 5% CO2 for 24 h. After exposure to different NP formulations (ASODN1-3, ASODN1-3-NP, SODN, NP and DMEM) or untreated as control for 24 h at 37, cell viability was evaluated by MTT assay (Yes Service Biotech, Inc.), and the absorbance was read on a micro-plate reader at 540 nm. The survival percentages were calculated as: Survival %=(A540 nm for treated cells/A540 nm for control cells) × 100%. Each assay was conducted in triplicate. Finally, concentration-viability curves were made and IC50 values were calculated.
Analysis of cell cycle
SHG44 glioma cells were seeded into 6-well culture plates at a density of 1 × 105 cells/well and cultured in DMEM medium supplemented with 10% FBS at 37°C in 5% CO2 for 24 h. The cells were treated with SODN-NP, ASODN or ASODN-NP for 48 h. The cells were collected and washed by PBS, and then cell cycle distribution was analyzed by flow cytometry.
RT-PCR
Total RNA was extracted from different groups of SHG44 cells using TRIzol reagent (Invitrogen, USA) following the manufacturer’s protocol. cDNA was synthesized by reverse transcription using RT kit (Promega, Madsion, WI, USA) following the manufacturer’s protocol. PCR was performed using the primers as follows: hTERT 5’- GTTTGGAAGAACCCCACAT-TT-3, and 5’- CGAGTCAGCTTGAGCAGGAAT-3’; β-actin 5’- TGACGTGGACTCCGCAAAG-3’ and 5’- CTGGAAGGTGGACAGCGAGG-3’. The amplification condition was as follows: 94°C for 4 min; 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec (35 cycles). The relative mRNA levels of hTERT were normalized to GAPDH and calculated by 2-ΔΔCt method.
Immunocytochemical analysis
SHG44 cells were treated by different NP. The cells were fixed by 3% paraformaldehyde for 30 min and washed by PBS twice. The telomerase protein was detected by EnVision Kit (DAKO) according to the manufacturer’s instructions.
Data analysis
Data analysis was performed by using SPSS 12.0 software. The results were expressed as means ± standard error (SE). ANOVA was used for the comparison among the groups. Statistical significance was set at P<0.05.
Results
Characterization of NP and SODN-NP/ASODN-N-NP
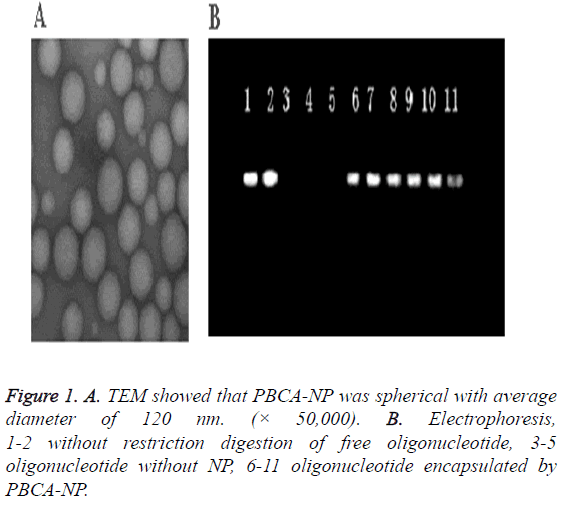
The nanoparticles were discrete and uniform spheres with average diameter of 120 nm (Figure 1A). Its size distribution was narrow. Its drug loading and entrapment efficiency for ASODN was 71.17% and 95.20%, respectively, and its Zeta potential was +41.3 mV.
Agarose gel electrophoresis showed that the control sample without restriction in the positive direction were free ODN, but we found no band in the positive direction of strip for ODN-NP, which indicated the protection effect of nanoparticles for oligonucleotide (Figure 1B).
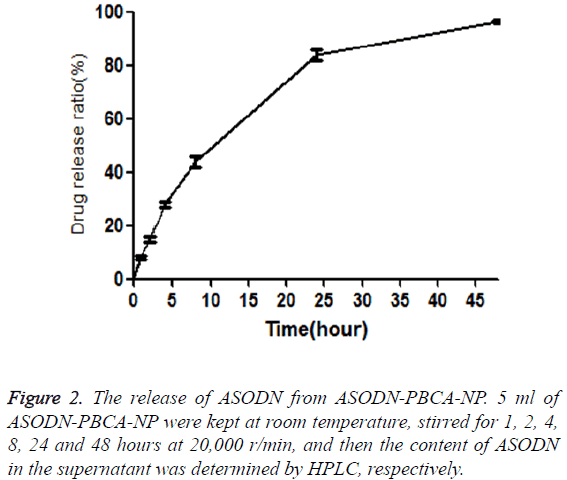
Cumulative release of ASODN from ASODN-NP was analyzed by drawing release curve. As shown in Figure 2, ASODN release increased with the increase of the time. Furthermore, the freeze dried SODN-NP/ASODN-NP powder were kept for three months under normal temperature and then dissolved in distilled water. The drug loading ratio (DL) and encapsulation efficiency (EE) were calculated, and they showed no significant difference compared to those of freshly prepared SODN-NP/ASODN-NP, indicating the stability of ASODN-NP.
ASODN-NP inhibited SHG44 cell viability
MTT assay showed that 48 hours after transfection, there was no significant difference in SHG44 viability among NP, SODN-NP and ASODN groups. Compared with control group, ASODN-NP exhibited significant inhibitory effect on SHG44 cell viability (P<0.01).
ASODN-NP inhibited SHG44 cell cycle progression
Flow cytometry analysis showed that 48 hours after transfection, there was no significant difference in cell cycle progression among NP, SODN-NP and ASODN group. However, the percentages of cells at G0/G1 and G2/M phases increased significantly while the percentages of cells at S phase decreased significantly in ASODN-NP group compared with other groups (P<0.01, Table 1).
| Group Percentage of cells in each phase (%) |
| G0/G1 S G2/M |
| Control (DMEM) 34.2 ± 1.4 60.1 ± 0.3 5.6 ± 0.1 |
| NP 35.3 ± 0.8 57.7 ± 1.6 6.9 ± 0.3 |
| SODN-NP 33.7 ± 1.6 61.4 ± 1.7 4.8 ± 0.5 |
| ASODN1 36.4 ± 0.3 56.3 ± 0.8 7.1 ± 0.4 |
| ASODN2 33.8 ± 1.2 62.2 ± 1.1 3.3±0.2 |
| ASODN3 36.9 ±1.9 55.8 ± 0.7 6.9 ± 0.6 |
| ASODN1-NP 62.8 ± 0.5 24.5 ± 0.6 12.5 ± 0.7 |
| ASODN2-NP 60.6 ± 2.2 27.3 ± 2.1 12.3 ± 0.3 |
| ASODN3-NP 63.4 ± 1.1 25.7 ± 1.4 10.1 ± 0.2 |
Table 1: Flow cytometry analysis of cell cycle in each group.
ASODN-NP inhibited hTERT mRNA expression in SHG44 cells
The level of hTERT mRNA in control group, NP group, SODN group, SODN-NP and ASODN-NP1-3 groups were 2.23 ± 0.12, 2.31 ± 0.14, 2.26 ± 0.19, 2.33 ± 0.16, 1.15 ± 0.13, 1.17 ± 0.21 and 1.08 ± 0.32 respectively. hTERT mRNA expression in SHG44 cells treated by ASODN-NP1-3 was significantly lower than that in control groups (P<0.05).
ASODN-NP inhibited telomerase protein expression in SHG44 cells
Telomerase protein expression was detected by immunocytochemical analysis. The results showed that the positive ratio of blank control group, NP group and SODN-NP were 94.6 ± 1.3%, 93.2 ± 3.1% and 93.7 ± 2.6%, respectively, showing no significant difference among three groups (P>0.05). However, positive ratio of ASODN1-NP, ASODN2- NP and ASODN3-NP group was 53.1 ± 1.8%, 56.7 ± 1.5% and 59.6 ± 4.3%, respectively, showing significant difference compared with blank control group (P<0.05).
Discussion
Telomerase is a reverse transcriptase enzyme which maintains telomeres by adding telomeric TTAGGG repeats to the ends of chromosomes [7]. High expression of telomerase is believed to be an essential step during malignant tumor progression [8,9]. Furthermore, high telomerase activity is usually associated with high tumor aggressiveness. It has been found that more than 50% gliomas have high telomerase activity, and the level of hTERT mRNA was predictive of glioma patient survival. Antisense treatment induced a decrease in hTERT mRNA expression, telomerase activity, cell viability, and an increase in apoptosis [10]. However, antisense oligonucleotide is difficult to penetrate cell membranes and blood-brain barrier, and prone to be degraded by endonuclease in the blood or CSF, which limits the use of antisense oligonucleotides in the clinic.
In this study, ASODN encapsulated in PBCA-NP was protected from the degradation by the nuclease. Maksimenko et al. reported that antisense oligonucleotides against EWSFli- 1 oncogene inhibited the growth of EWS-Fli-1 dependent Ewing’s tumor grafted to nude mice with high specificity after being encapsulated by nanocapsules or nanospheres [11]. The main advantages of using nanoparticles for brain drug targeting include their ability to deliver drugs to the brain without changing their original characteristics, decrease drug escape in the brain and reduce peripheral toxicity. It was found that oligonucleotides encapsulated into polyisohexylcyanoacrylate nanoparticles are protected from nuclease attack and increase cellular uptake of oligonucleotides by an endocytic/phagocytic pathway [12].
In this study, after being encapsulated into nanoparticles, the stability of ASODN increased significantly, which was confirmed by the electrophoretic mobility shift assay. After SHG44 glioma cells were treated with ASODN-NP, we found reduced hTERT mRNA and reduced telomerase protein expression levels, reduced cell viability, increased percentages of cells at G0/G1 and G2/M phases and decreased percentages of cells at S phase compared to control groups.
In conclusion, nanoparticles obtained by emulsion polymerization method are steady with high drug loading and entrapment efficiency of oligodeoxynucletide. Polybutylcyanoacrylate nanoparticles loaded with antisense oligodeoxynucleotide of hTERT can enter into glioma cells, block the expression of hTERT, and inhibit cell viability. Thus they are promising agent for the treatment of glioma.
Acknowledgements
This study was supported by National Nature Science Foundation of China (No. 81401500, 81271554, 81472739), Science Foundation of Jiangsu Province (No. BK20140298), and Project Fund by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY. Primary brain tumours in adults. Lancet 2003; 361: 323-331.
- Stan AC, Casares S, Radu D, Walter GF, Brumeanu TD. Doxorubicin induced cell death in highly invasive human gliomas. Anticancer Res 1999; 19: 941-950.
- Sugita Y, Nakashima A, Kato S, Sakata K, Morimatsu M, Shigemori M. Telomerase activity in gliomas with the use of non-radioisotopic and semi-quantitative procedure for terminal repeat amplification protocol. Oncol Rep 2000; 7: 1087-92.
- Chaudhari KR, Ukawala M, Manjappa AS, Kumar A, Mundada PK, Mishra AK, Mathur R, Mönkkönen J, Murthy RS. Opsonization, biodistribution, cellular uptake and apoptosis study of PEGylated PBCA nanoparticle as potential drug delivery carrier. Pharm Res 2012; 29: 53-68.
- Zhang Y, Yu J, Zhang L, Cai J, Cai D, Lv C. Enhanced anti-tumor effects of doxorubicin on glioma by entrapping in polybutylcyanoacrylate nanoparticles. Tumour Biol 2016; 37: 2703-2708.
- Ebrahimi Shahmabadi H, Movahedi F, Koohi Moftakhari Esfahani M, Alavi SE, Eslamifar A, Mohammadi Anaraki G, Akbarzadeh A. Efficacy of Cisplatin-loaded polybutyl cyanoacrylate nanoparticles on the glioblastoma. Tumor Biology 2014; 35: 4799-4806.
- Chen YC, Huang FC, Lin JJ. The addition of a spin column step in the telomeric repeat application protocol removes telomerase inhibitors. Anal Biochem 2015; 478: 49-51.
- Dirk Hockemeyer. Elucidating Telomerase Function in Human Tumor and Stem Cell. FASEB J 2015; 29: Supplement 84.1.
- Gupta R, Dong Y, Solomon PD, Wettersten HI, Cheng CJ, Min JN, Henson J, Dogra SK, Hwang SH, Hammock BD, Zhu LJ, Reddel RR, Saltzman WM, WeissRH, Chang S, Green MR, Wajapeyee N. Synergistic tumor suppression by combined inhibition of telomerase and CDKN1A. PNAS 2014; 111: E3062–E3071.
- Tao Z, Chen S, Wu Z, Xiao B. Targeted therapy of human laryngeal squamous cell carcinoma in vitro by antisense oligonucleotides directed against telomerase reverse transcriptase mRNA, J Laryngol Otol 2005; 119: 92-96.
- Maksimenko A, Malvy C, Lambert G, Bertrand JR, Fattal E, Maccario J, Couvreur P. Oligonucleotides Targeted Against a Junction Oncogene Are Made Efficient by Nanotechnologies. Pharm Res 2003; 20: 1565-1567.
- Yezhelyev MV, Qi L, O'Regan RM, Nie S, Gao X. Proton-Sponge Coated Quantum Dots for siRNA Delivery and Intracellular Imaging. J Am Chem Soc 2008; 130: 9006-9012.