ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 14
Role of GATA-3 and mir-135a in the exercise induced immunosuppression
1Department of Chemistry, Qilu Medical University, Zibo, P.R.China
2Department of Exercise Physiology, Shandong Research Center of Sport Science, Jinan 250102, P.R.China
3Key Laboratory of Biomedical Engineering and Technology of Shandong High School, Qilu Medical University, Zibo, P.R.China
#These authors contributed equally to this work
- *Corresponding Author:
- Qinglu Wang
Key Laboratory of Biomedical Engineering and Technology of Shandong High School
Qilu Medical University, Zibo, P.R.China
Accepted date: June 20, 2017
Aims: This study is to investigate the role of GATA-3 and miR-135a in immunosuppression induced by exercise.
Methods: Rats received load exercise to induce immunosuppression. ELISA was used to detect the change of testosterone, cortisol, IFN-γ and IL-4. The spleen index and thymus index was calculated. The qRT-PCR and Western Blot were used to detect the mRNA and protein change of GATA-3 in spleen tissue. The upstream microRNA for GATA-3 was predicted using bioinformatics. The miRNA-135a expression change was detected in spleen tissue using qRT-PCR. The dual luciferase assay was used to identify whether GATA-3 was the target gene for miRNA-135a.
Results: Compared with control group, testosterone and IFN-γ significantly decreased while IL-4 significantly increased in model group (P<0.05). The spleen index significantly decreased in model group (p<0.05). The mRNA and protein of GATA-3 significantly increased in spleen tissue and miRNA-135a significantly decreased in model group (P<0.05). While there were no significant difference of thymus index between model and control group. Dual luciferase assay showed GATA-3 was the target gene for miRNA-135a.
Conclusions: The significantly increased mRNA and protein of GATA-3 may be related to the down regulation of miRNA-135a. GATA-3 and mir-135a may play a role in the progression of exercise-induced immunosuppression.
Keywords
miRNA-135a, GATA-3, Th1/Th2, Exercise induced immunosuppression.
Introduction
The fatigue induced by high-intensity exercise may lead to immunosuppression [1]. The exercise induced immunosuppression and increased susceptibility to infectious diseases often result in a decline or stagnation of athlete’s performances [2]. Currently, the most important indicators of exercise induced fatigue are blood testosterone and corticosterone levels [3]. The imbalance of T-lymphocyte differentiation, especially the helper T cells (Th cells), is an important factor in exercise induced immunosuppression [4]. Th1 and Th2 are the two subtypes of Th cells. Excessive exercise leads to increased Th2 and decreased Th1/Th2 ratio [5,6]. GATA binding protein 3 (GTA3) is a Th2-type specific transcription factor that is only expressed in Th2 cells and is essential in the Th2 differentiation [7,8]. Cytokine IL-4 is produced by Th2 cells. GATA-3 is also the main factor to induce the differentiation of Th0 to Th2 cells. GATA-3 has strong antagonizing effect for IFN-γ produced by Th1. It is shown that IL-4 mRNA changed significantly after exercise [9,10]. Therefore, GATA-3 is of great importance in the Th2 regulation.
The miRNA is a class of small non-coding RNA with length of 18-22 nt that is ubiquitous in eukaryotes and can regulate the protein expression at mRNA level [11-13]. In the exercise induced immunosuppression, the expression levels of many miRNAs and proteins are changed [14]. The miR-135a is involved in the regulation of many diseases. For example, serum mir-135a can be used as a supplementary diagnostic biomarker for colorectal cancer [15]. mir-135a can be used as biomarker to detect blood lipids and inflammatory changes [16]. mir-135a can inhibit the calcification of senescent vascular endothelial cell by regulation of KLF4/STAT3 signalling pathway [17]. mir-135a may also influence the epithelial-mesenchymal transition process of lung cancer cells [18]. However, the relationship between miR-135a and GATA-3 is unclear.
In this study, qRT-PCR, Western blot, ELISA and dual luciferase assay were used to detect the cytokines, spleen index, and gene and protein expression in spleen in the exercise induced immunosuppression rat model.
Materials and Methods
Animals
The 40 Specific Pathogen Free (SPF) grade healthy Sprague Dawley male rats with weight of 200-250 g (Chongqing Xin Teng Bill animals Sales Co., Ltd. approval number SCXK (Chongqing) 2014-0008) were used. Before experiment, all rats received one-week adaptive feeding, with free access to food and drinking water. All animal procedures were in accordance with 3R (“Reduction”, “Replacement”, and “Refinement”) animal welfare principles. All animal experiments were conducted according to the ethical guidelines of Qilu Medical University.
Reagents and instruments
The following reagents and instruments were used in this study. The MiRcute MiRNA Isolation Kit (TIANGEN, Beijing), miRcute miRNA cDNA First Strand Synthesis Kit (TIANGEN, Beijing), MiRcute MiRNA Quantitative Fluorescence Detection Kit (FP401, TIANGEN, Beijing), Super Real PreMix (SYBR Green, FP204, TIANGEN, Beijing), TIANScript II cDNA First Strand Synthesis Kit (KR107, TIANGEN, Beijing), qRT-PCR instrument (Bio-Rad fluorescence quantitative PCR-iQ5), GATA-3 primary antibody (ab199428, rabbit anti-human, abcam, USA), Trizol Reagent (10606ES60, Yisheng Biology, Shanghai), β-actin antibody (rabbit anti-mouse, ab129348, abcam, USA), secondary antibody (ab6721, goat anti-rabbit, abcam, USA), BCA Protein Assay Reagent Kit (RTP7102, Zhongke Ruitai, Beijing), Serum RNA Extraction miRNeasy Serum/Plasma kit (50) (JL217184, Jianlun Biology, Guangzhou), IL-4 ELISA Kit (ab100770, abcam, USA), IFN-γ ELISA kit (ab 113349, abcam, USA), testosterone Kit (No: jn-ELA-5850, Shanghai Jining Biological Technology Co., Ltd.), rat cortisol ELISA Assay kit (No: GD-SZ2110, Shanghai Guduo Biological Technology Co., Ltd.), image lab software 3.0 (Bio-Rad Laboratories, Hercules, CA, USA), and ECL Luminescent Liquid (ab65623, abcam, USA). All the plasmids/agomiR were designed and synthesized by Shanghai Biological Technology Co., Ltd.
The establishment of exercise induced immunosuppression rat model
The rats were randomly divided into control group and model group, with 20 rats in each group. The control group was in normal living conditions without load exercise. In model group, all rats had one-week adaptive exercise (speed of 15 m/ min; gradient of 0°C; time of 10 min/d) before formal training.
Then, rats received load exercise. The load exercise training schedule was shown in Table 1.
| Week | Speed (m/min) | Slope (%) | Time (min) |
|---|---|---|---|
| 1 | 15 | 2 | 40 |
| 2 | 20 | 10 | 60 |
| 3 | 25 | 10 | 90 |
| 4 | 30 | 5 | 120 |
| 5 | 30 | 5 | 120 |
| 6 | 30 | 8 | 120 |
| 7 | 35 | 10 | Exhausted |
| 8 | 35 | 15 | Exhausted |
| 9 | 35 | 15 | Exhausted |
Table 1: The load exercise training schedule.
Specimen collection
After 9 weeks training, rats were anesthetized by the 1% sodium pentobarbital (1 ml/100 g). The 5 ml of abdominal aortic blood was collected. Then serum was isolated and stored at -80°C. The spleen and thymus tissues were collected and kept at -80°C.
ELISA
The serum levels of testosterone, cortisol, IFN-γ and IL-4 were detected with ELISA kits, respectively, according to the instructions provided by the kits. Briefly, 50 μl standard samples of different concentrations were added to each standard well. The 10 μl serum along with 40 μl sample dilution solution was added to each test well. Except for the blank control, a 100 μl of Horseradish Peroxidase (HRP) labelled detection antibody was added to each well, and incubated for 1 h. After 5 times washing, a 50 μl substrate A and B were added in each well and were incubated for 15 min at 37°C. The OD value was measured at 450 nm wavelength within 15 min after adding the 50 μl stop solution.
Spleen index and thymus index
All rats were weighed and sacrificed. Spleens were removed and weighed. The spleen and thymus index were defined as organ weight per 10 g body weight, calculated as thymus weight (mg)/body weight (g) × 103.
Quantitative RT-PCR (q-RT-PCR)
Total RNAs were extracted from spleen tissues by Trizol using phenol-chloroform extraction method. The RNA integrity was examined by gel electrophoresis, and RNA purity was assessed by 260/280 ratio by spectrophotometry. RNA was reverse transcribed into cDNA. The primer sequences of GATA-3, GAPDH, miR-135a and U6 for qRT-PCR were shown in Table 2. GAPDH and U6 were used as internal control for GATA-3 and miR-135a, respectively. The PCR procedure for GATA-3 was as follows: pre-denaturation for 10 min at 95°C, followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 20 s at 55°C, and extending for 30 s at 72°C. The reaction conditions for miR-135a were pre-denaturation at 95°C for 5 min and 40 cycles of denaturation at 95°C for 20 s and annealing at 60°C for 30 s. The relative levels of GATA-3 and miR-135a were calculated using 2-ΔΔCt method.
| Primer | Sequences |
|---|---|
| GATA-3 | Forward 5'- AAGAGTGCCTCAAGTATCAG -3' |
| Reverse 5'- GCGGATAGGTGGTAATGG-3' | |
| GAPDH | Forward 5'- CCCTCAATGACCACTTTGTG-3' |
| Reverse 5'- GGTTTGAGGGCTCTTACTCCT-3' | |
| miR-135a | Forward 5’-GCGCCGTATGGCTTTTTATTCCTA-3’ |
| Reverse 5’-TGCAGAGATGTCCAGTCAGC-3’ | |
| U6 | Forward 5’-AACGCTTCACGAATTTGCGT-3’ |
| Reverse 5’-CTCGCTTCGGCAGCACA-3’ |
Table 2: Primer sequences.
Western blot
Total protein was extracted by protein lysate and its concentration was measured using BCA protein assay kit. Then, proteins were separated by 10% SDS-polyacrylamide electrophoresis and transferred to PVDF membrane. The membrane was blocked by 5% skim milk for 1 h. After blocking, the primary antibodies of GATA-3 and β-actin were added and incubated overnight at 4°C. Then, the secondary antibody was added and incubated at room temperature for 1 h. The membrane was developed in ECL luminescent liquid and the developed film was scanned in gel imaging system using image lab software. The relative expression level of GATA-3 was calculated based on the grey value of β-actin.
The bioinformatics prediction of the regulatory upstream miRNA for GATA-3
Bioinformatics prediction was used to identify the upstream miRNA for GATA-3. The following gene prediction software’s were used for bioinformatics prediction: miRanda (http://www.microma.org/rnicroma/home.do), TargetSean (www.targetscan.org), PiTa (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and PICTA (http://pictar.mdc-berlin.de/). The miR-135a was found to be the potential upstream gene of GATA-3.
Dual luciferase assay
The wild type and mutant type of miR-135a binding sequence in the 3'-UTR region of GATA-3 gene was constructed by in vitro chemical synthesis. The cleavage sites of Spe-1 and Hind III were respectively added on both ends. The two DNA fragments were cloned into pMIR-REPORT luciferase plasmids. Using liposome method, the plasmids with wild type 3'-UTR and mutant type 3'-UTR sequences were transfected into 293 T cells, respectively. Then, agomiR-135a (100 nM) was transfected and incubated for 24 h. The fluorescence values were measured using GloMax 20/20 luminometer. The Renilla fluorescent activity was used as internal control, and all procedures were strictly in accordance with the dual luciferase assay kit instructions.
Statistical analysis
All data were processed by SPSS18.0 (SPSS Inc, Chicago, IL, USA). All data were presented as mean ± standard deviation. All data preformed normality test. Multiple groups of measurement data were analysed using one-way ANOVA. Fisher’s Least Significant Difference (LSD) and Student- Newman-Keuls (SNK) method was used when there was homogeneity of variance; otherwise, Tamhane’s T2 or Dunnett’s T3 method was used. P<0.05 was considered statistically significant.
Results
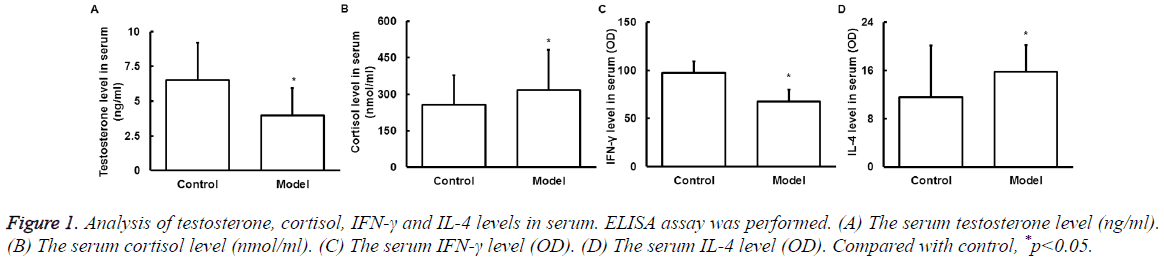
Serum testosterone, cortisol, IFN-γ and IL-4 levels
ELISA was used to detect serum testosterone, cortisol, IFN-γ, and IL-4 level changes after exercise. It is found that compared with control, the testosterone (Figure 1A) decreased significantly and cortisol (Figure 1B) increased significantly after exercise (P<0.05), indicating the overload fatigue. The IFN-γ (Figure 1C) was significantly decreased and IL-4 (Figure 1D) was significantly increased (P<0.05), indicating the Th1/Th2 imbalance and Th2 drift.
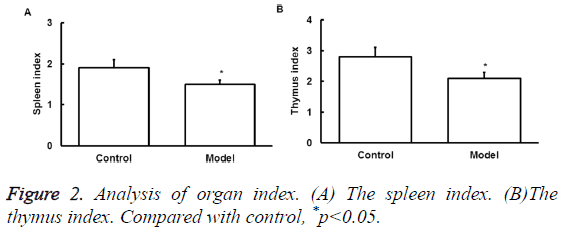
Effect of exercise on the immune organ index
To determine the effect of exercise on the immune organ index, spleen and thymus index were assessed. Thymus and spleen are two important immune organs that can represent the overall immune activity. Results showed that excessive exercise reduced spleen index (Figure 2A). Thymus index was also reduced (Figure 2B). This data indicates the immunosuppression of exercise in rat model.
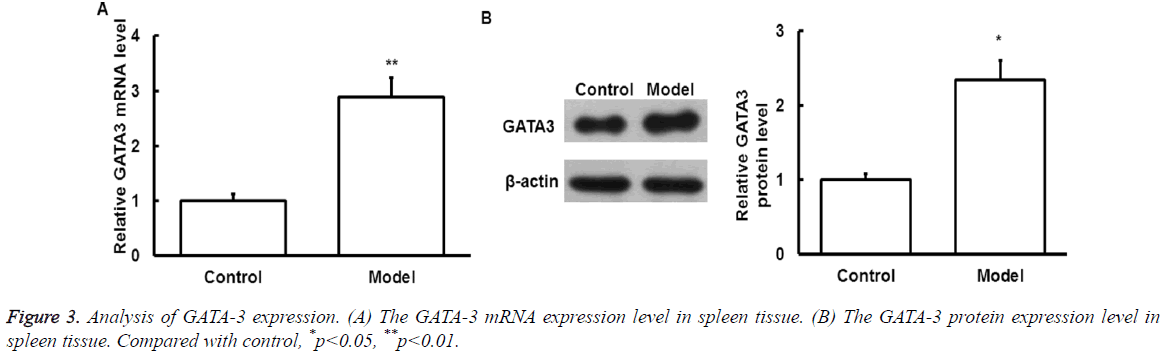
GATA-3 mRNA and protein expression
To detect the GATA-3 mRNA and protein expression level after exercise, qRT-PCR and Western Blot was performed, respectively.
As shown in Figure 3, mRNA (Figure 3A) and protein (Figure 3B) of GATA-3 in spleen samples of model group significantly increased compared with that of control group (p<0.05).
The above results suggest that GATA-3 expression level is increased after exercise.
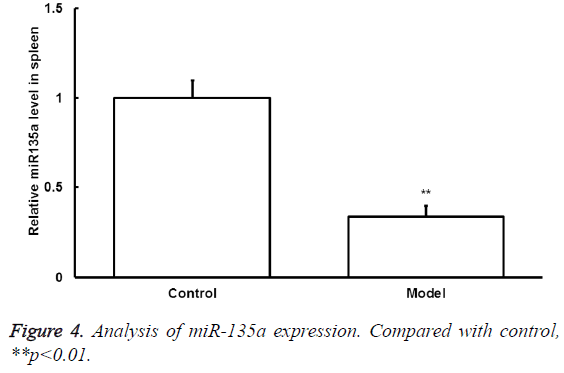
The miRNA-135a expression
To detect the changes of miRNA-135a expression after exercise, qRT-PCR was conducted. The mRNA-135a expression level in spleen samples of model group significantly decreased compared with that of control group (p<0.05) (Figure 4).
Dual luciferase assay
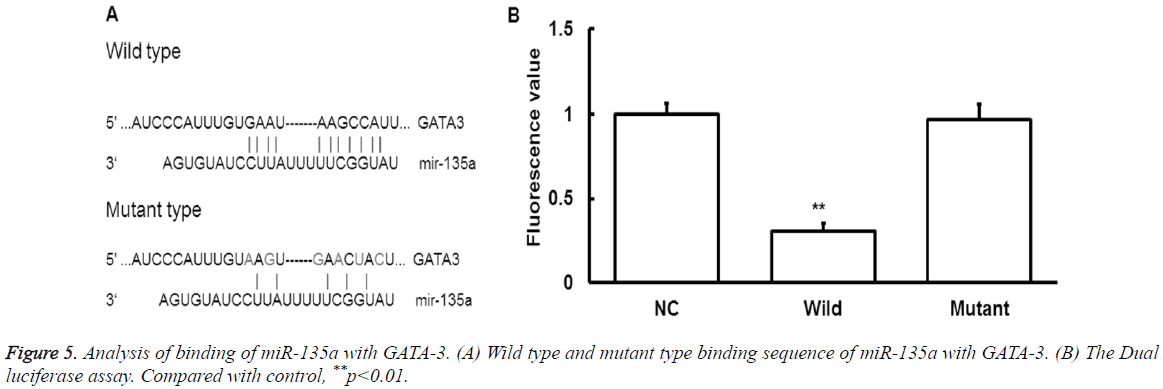
To find the upstream regulatory miRNA of GATA-3, bioinformatics prediction was performed. The miR-135a was found to be the potential upstream gene of GATA-3. The wild type and mutant type of binding sequence were shown in Figure 5A.
To determine whether miR-135 could directly target GATA-3, dual luciferase assay was performed. It showed that the fluorescence values significantly decreased after the cotransfection of agomiR-135a and pmiR-report (P<0.05) (Figure 5B).
While there were no statistically significant differences in fluorescence values compared with the mutant group (P>0.05). It indicates that miR-135a can directly bind with GATA-3 at the 3'-UTR region to regulate its expression. Together, these results indicate that miRNA-135a may play a regulatory role in the pathological process of exercise-induced immunosuppression through GATA-3.
Discussion
In this study, we investigated the changes of serum cytokines, the spleen index, the thymus index, GATA-3 expression and miRNA-135a expression after load exercise. The potential mechanism was discussed.
The exercise induced immunosuppression refers to the declined immune function after excessive exercise, and it remains an unsolved issue in sports medicine. Recently, the Th1/Th2 imbalance linked the post exercise neuroendocrineimmunity changes to cytokines and this may be the underlying mechanism of exercise induced immunosuppression [19,20].
Th1 and Th2 are the two subtypes of helper T cells. Th1 cells secrete IL-2 and IFN-γ while Th2 cells secrete IL-4 and IL-5. Th1/Th2 is in a balance in human body [21]. Th1/Th2 balance can be used to evaluate the dynamic changes of cellular immunity and humoral immunity during exercise, and the impact of exercise on Th1/Th2 immune imbalance has been reported [22-25]. Th1/Th2 imbalance can be reflected by serum cytokines. Th1 specifically produces IFN-γ but not IL-4; while on the other hand, Th2 specifically produces IL-4 but not IFN-γ. Therefore, IFN-γ and IL-4 are often used to evaluate the Th1/Th2 balance [26]. In our study, testosterone decreased and cortisol increased after exercise, indicating that there is the excessive exercise induced fatigue. The spleen and thymus index both decreased after exercise, indicating the immunosuppression. The IFN-γ significantly decreased and IL-4 increased, indicating the imbalanced Th1/Th2 and Th2 drift. Therefore, these results suggest that the exercise may induce immunosuppression through imbalanced Th1/Th2.
The transcription factor of IL-4 is GATA-3, which promotes IL-4 gene transcription, inhibits IFN-γ expression, regulates Th2 cell differentiation and cytokine expressions, as well as promotes Th1 transfer to Th2 [27]. The expression level of GATA-3 is low in naive CD4+ T cells [28]. However, the expression level of GATA-3 increases when naive T cells differentiate into Th2 and decreases when naive T cells differentiate into Th1 [29]. GATA-3 cannot be detected in mature Th1 cells, therefore, GATA-3 is specifically expressed in Th2 cells [29]. It is generally believed that GATA directly activate IL-4 gene through two GATA-3 binding sites [30]. In our study, it indicates that mRNA and protein level of GATA-3 significantly increased in rat model group compared with that of control group, which may lead to IL-4 protein up-regulation. This also suggests that the Th1/Th2 imbalance drifts toward Th2 increase.
In addition, microRNA, the upstream regulator of GATA-3, was investigated. The microRNA is a class of endogenous, small, non-coding RNA miRNA [31,32]. The miRNAs are vital regulatory factors in the disease development, physiology and pathogenesis [33,34]. The upstream regulatory gene for GATA-3 was predicted by bioinformatics and miR-135a was found to be one of the potential upstream genes that might regulate GATA-3 expression. The dual luciferase assay further confirmed that miR-135a could directly bind with the 3’UTR region of GATA-3, suggesting that miR-135a could directly regulate GATA-3 expression.
In conclusion, the mRNA and protein expression of GATA-3 in the spleen tissues was significantly increased whereas miR-135a level was significantly decreased in exerciseinduced immunosuppression model. GATA-3 was one of the direct targets of miR-135a. GATA-3 and miR-135a may participate in immunosuppression induced by exercise.
Acknowledgements
This work was supported by grants from the Natural Scientific Foundation of Shandong Province, China (ZR2014CM046, ZR2010CQ031 and ZR2015CL019) and Collaborative Innovation Center of Chinese medicine antivirus in Shandong University of Traditional Chinese Medicine (XTCX2014B01-07).
Conflict of Interest
The authors declare no conflict of interests.
References
- Penkman MA, Field CJ, Sellar CM, Harber VJ, Bell GJ. Effect of hydration status on high-intensity rowing performance and immune function. Int J Sports Physiol Perform 2008; 3: 531-546.
- Ahmed M, Henson DA, Sanderson MC, Nieman DC, Gillitt ND, Lila MA. The protective effects of a polyphenol-enriched protein powder on exercise-induced susceptibility to virus infection. Phytother Res 2014; 28: 1829-1836.
- Shin S, Yeon S, Park D, Oh J, Kang H, Kim S, Joo SS, Lim WT, Lee JY, Choi KC, Kim KY, Kim SU, Kim JC, Kim YB. Silk amino acids improve physical stamina and male reproductive function of mice. Biol Pharm Bull 2010; 33: 273-278.
- Song QH, Xu RM, Zhang QH, Shen GQ, Ma M, Zhao XP, Guo YH, Wang Y. Glutamine supplementation and immune function during heavy load training. Int J Clin Pharmacol Ther 2015; 53: 372-376.
- de Lastic AL, Rodi M, Mouzaki A. Effect of dendritic cell state and antigen-presentation conditions on resulting T-cell phenotypes and Th cytokine profiles. Immunobiol 2016; 221: 862-870.
- Lancaster GI, Halson SL, Khan Q, Drysdale P, Wallace F, Jeukendrup AE, Drayson MT, Gleeson M. Effects of acute exhaustive exercise and chronic exercise training on type 1 and type 2 T lymphocytes. Exerc Immunol Rev 2004; 10: 91-106.
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000; 100: 655-669.
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA 2001; 98: 15137-15142.
- Lametschwandtner G, Biedermann T, Schwarzler C, Gunther C, Kund J, Fassl S, Hinteregger S, Carballido-PN, Szabo SJ, Glimcher LH, Carballido JM. Sustained T-bet expression confers polarized human TH2 cells with TH1-like cytokine production and migratory capacities. J Allergy Clin Immunol 2004; 113: 987-994.
- Mueller O, Villiger B, O’Callaghan B, Simon HU. Immunological effects of competitive versus recreational sports in cross-country skiing. Int J Sports Med 2001; 22: 52-59.
- Graziano A, Lo Monte G, Piva I, Caserta D, Karner M, Engl B, Marci R. Diagnostic findings in adenomyosis: a pictorial review on the major concerns. Eur Rev Med Pharmacol Sci 2015; 19: 1146-1154.
- Jia W, Wu Y, Zhang Q, Gao GE, Zhang C, Xiang Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol Clin Oncol 2015; 3: 851-858.
- Jiang XI, Luo Y, Zhao S, Chen Q, Jiang C, Dai Y, Chen Y, Cao Z. Clinical significance and expression of microRNA in diabetic patients with erectile dysfunction. Exp Ther Med 2015; 10: 213-218.
- Liu G, Detloff MR, Miller KN, Santi L, Houle JD. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Exp Neurol 2012; 233: 447-456.
- Wang Q, Zhang H, Shen X, Ju S. Serum microRNA-135a-5p as an auxiliary diagnostic biomarker for colorectal cancer. Ann Clin Biochem 2016; 28.
- Desgagne V, Guay SP, Guerin R, Corbin F, Couture P, Lamarche B, Bouchard L. Variations in HDL-carried miR-223 and miR-135a concentrations after consumption of dietary transfat are associated with changes in blood lipid and inflammatory markers in healthy men - an exploratory study. Epigenetics 2016; 11: 438-448.
- Lin L, He Y, Xi BL, Zheng HC, Chen Q, Li J, Hu Y, Ye MH, Chen P, Qu Y. MiR-135a Suppresses Calcification in Senescent VSMCs by Regulating KLF4/STAT3 Pathway. Curr Vasc Pharmacol 2016; 14: 211-218.
- Shi H, Ji Y, Zhang D, Liu Y, Fang P. MiR-135a inhibits migration and invasion and regulates EMT-related marker genes by targeting KLF8 in lung cancer cells. Biochem Biophys Res Commun 2015; 465: 125-130.
- Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O’Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med 2000; 192: 105-115.
- Skapenko A, Leipe J, Niesner U, Devriendt K, Beetz R, Radbruch A, Kalden JR, Lipsky PE, Schulze-Koops H. GATA-3 in human T cell helper type 2 development. J Exp Med 2004; 199: 423-428.
- Romagnani S. Human TH1 and TH2 subsets: doubt no more. Immunol 1991; 12: 256-257.
- Lin Y, Zhou X, Guo W, Li Q, Pan X, Bao Y, He M, Zhu B, Lin X, Jin L, Yao R. RhIL-11 treatment normalized Th1/Th2 and T-bet/GATA-3 imbalance in in human immune thrombocytopenic purpura (ITP). Int Immunopharmacol 2016; 38: 40-44.
- Feng Y, Tian J, Xie HQ, She J, Xu SL, Xu T, Tian W, Fu H, Li S, Tao W, Wang L, Chen Y, Zhang S, Zhang W, Guo TL, Zhao B. effects of acute low-dose exposure to the chlorinated flame retardant dechlorane 602 and Th1 and Th2 immune responses in adult male mice. Environ Health Perspect 2016; 15.
- Gao M, Wu Y, Li GQ. Regulating effect of moxibustion pretreatment on Th1/Th2 imbalance of athletes during course of heavy load training. Zhongguo Zhen Jiu 2011; 31: 247-251.
- Ru W, Peijie C. Modulation of NKT cells and Th1/Th2 imbalance after alpha-GalCer treatment in progressive load-trained rats. Int J Biol Sci 2009; 5: 338-343.
- Kutukculer N, Azarsiz E, Aksu G, Karaca NE. CD4+ CD25+Foxp3+ T regulatory cells, Th1 (CCR5, IL-2, IFN-gamma) and Th2 (CCR4, IL-4, Il-13) type chemokine receptors and intracellular cytokines in children with common variable immunodeficiency. Int J Immunopathol Pharmacol 2016; 29: 241-251.
- Hosokawa H, Tanaka T, Endo Y, Kato M, Shinoda K, Suzuki A, Motohashi S, Matsumoto M, Nakayama KI, Nakayama T. Akt1-mediated GATA-3 phosphorylation controls the repression of IFN gamma in memory-type Th2 cells. Nat Commun 2016; 7: 11289.
- Baraut J, Farge D, Jean-LF, Masse I, Grigore EI, Arruda LC, Lamartine J, Verrecchia F, Michel L. Transforming growth factor-beta increases interleukin-13 synthesis via GATA-3 transcription factor in T-lymphocytes from patients with systemic sclerosis. Arthritis Res Ther 2015; 17: 196.
- Hosokawa H, Tanaka T, Endo Y, Kato M, Shinoda K, Suzuki A, Motohashi S, Matsumoto M, Nakayama KI, Nakayama T. Akt1-mediated GATA-3 phosphorylation controls the repression of IFN gamma in memory-type Th2 cells. Nat Commun 2016; 7: 11289.
- Seki N, Miyazaki M, Suzuki W, Hayashi K, Arima K, Myburgh E, Izuhara K, Brombacher F, Kubo M. IL-4-induced GATA-3 expression is a time-restricted instruction switch for Th2 cell differentiation. J Immunol 2004; 172: 6158-6166.
- Inoue K. MicroRNA functions in animal development. Tanpakushitsu Kakusan Koso 2007; 52: 197-204.
- Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn 2007; 236: 572-580.
- Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao R, Cui L. The tumour suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol 2015; 46: 798-808.
- Lv ZC, Fan YS, Chen HB, Zhao DW. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol 2015; 36: 1619-1625.