ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 5
Roles of miR-16 in vascular endothelial injury in patients with coronary heart disease
Department of Emergency, Shanghai Ninth People's Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Shen Wu
Department of Emergency, Shanghai Ninth People's Hospital
School of Medicine, Shanghai Jiao Tong University, PR China
Accepted date: October 31, 2016
Objective: This study is to detect miR-16 expression in peripheral blood of patients with Coronary Heart Disease (CHD), and explore the role of miR-16 in vascular endothelial injury.
Methods: Peripheral blood was collected from coronary patients. QRT-PCR was applied to detect miR-16 expression and autophagy related 14 (AGT14) mRNA expressions. After transfection with miR-16 inhibitor in vitro, the biological functions of miR-16 were detected in endothelial cells by CCK-8, apoptosis assay, and cell cycle analysis. Bioinformatics methods predicted that ATG14 was one of target genes regulated by miR-16. Dual luciferase assay was used to validate whether miR-16 directly regulated ATG14. Expression changes of ATG14, Beclin1 and microtubule-associated protein 1 light chain 3 beta (LC3B) were detected by Western blot.
Results: miR-16 expression was significantly increased in blood in CHD patients, and miR-16 expression was higher in patients with unstable angina than patients with stable angina (P<0.05), which indicated that miR-16 was related to the development of CHD. In vitro experimental results showed that miR-16 inhibited the proliferation and promoted apoptosis of endothelial cells significantly (P<0.05). Compared with control group, the percentage of cells in G1 phase in miR-16 inhibitor group significantly decreased (58.8 ± 3.51 vs. 40.7 ± 3.32) whereas the percentage of cells in S phase significantly increased (21.8 ± 1.62 vs. 32.7 ± 2.47) (P<0.05), indicating that miR-16 inhibited G1/S conversion of endothelial cells. The dual luciferase assay validated that miR-16 directly targeted ATG14. Western blot and qRT-PCR results demonstrated that expressions of Beclin1 and LC3B were repressed when miR-16 was over-expressed while were significantly increased after down-regulated miR-16. And, electronic microscopy showed that miR-16 inhibited the formation of autophagy bodies in endothelial cells.
Conclusion: MiR-16 expression was up-regulated in peripheral blood of patients with CHD. MiR-16 regulated ATG14 expression in endothelial cells. Thus, miR-16 may play a role in the development of vascular endothelial injury and CHD.
Keywords
MiR-16, Vascular endothelial injury, Autophagy related 14 (AGT14), Coronary heart disease (CHD)
Introduction
Coronary Heart Disease (CHD) is a type of serious disease, which frequently occurs in cardiovascular system and is closely related to the environment, autoimmune, and genetic factors [1-3]. The pathogenic processes of CHD are very long, and the chronic vascular endothelial injury is a major cause of this disease [4,5]. It is demonstrated that endothelial cells have endocrine, paracrine and autocrine ability and can secrete biological active substances to regulate vascular function, such as nitric oxide, prostacyclin I2, thromboxane A2, which play important roles in occurrence and development of CHD [6,7]. In addition, the basic pathological change of atherosclerosis is the injury of vascular endothelial cells, which is a risk factor for CHD [8,9]. However, the exact pathogenesis mechanisms are still unclear [10].
MiRNA is a class of small non-coding RNA with 18-22 nt in length, which can inhibit protein translation through binding to 3’ UTR of target mRNA and then regulate gene expression as an important post-transcriptional regulator [11]. Bioinformatics algorithm predicts that about 1/3 genes can be regulated by miRNA [12]. It is demonstrated that miRNA expression profile changes significantly in peripheral blood in patients with cardiovascular diseases, and some of them could be used as diagnosis markers, such as miR-1, miR-135, and miR-126 [13,14]. Mature miR-16 is processed from precursor miR-16-1 and miR-16-2. MiR-16-1 is located in one fragile site on 11q24, and miR-16-2 is located in 21q211 near Let-7c [10]. MiR-16 could inhibit tumor proliferation, metastasis and promote apoptosis and its expression is down-regulated in lung cancer, liver cancer, ovarian cancer, and pancreatic cancer, etc. [15-17]. miR-16 is also related to cardiovascular diseases. For example, miR-16 inhibits angiogenesis through downregulating expressions of VEGFR2 and FGFR1 [18]. The expression change of miR-16 can induce myocardial cell hypertrophy [19]. The role and mechanism of miR-16 in CHD are not clear.
In this study, we aim to detect the expression of miR-16 in peripheral blood in patients with CHD and explore its role in vascular endothelial cells.
Materials and Methods
Clinical data and sample collection
This study included 67 cases of CHD patients, which was consisted by 37 cases of Unstable Angina Pectoris (UAP) and 30 cases of Stable Angina Pectoris (SAP) from February 2013 to August 2014 in Department of Cardiology in Shanghai Ninth People's Hospital. Among all the patients, 31 cases were male and 36 cases were female, and the mean age was (63.2 ± 5.36) years old. For control, 30 healthy individuals (21 males and 9 females) were included from center of health examination with the mean age of (60.80 ± 4.5) years old. All the patients were diagnosed based on 2011 “ACC/AHA Unstable Angina (UA)/non-ST Segment Elevation Myocardial Infarction (NSTEMI) Guide" and standard treatment guidelines of chronic stable angina by the Chinese Medical Association. Totally 5 ml fasting blood was collected from each patient and each healthy individual within 24 h of admission. Prior written and informed consent were obtained from every patient and the study was approved by the ethics review board of Shanghai Ninth People's Hospital.
Human umbilical vein endothelial cell (HUVEC) culture
DMEM medium with 10% FBS was used to culture HUVEC cells under 37°C in 5% CO2. When cell confluence reached about 70%-90%, trypsin digestion method was used for cell passage, and the first 3-6th generation cells were used for experiments.
Quantitative real time PCR (qRT-PCR)
Total RNAs of peripheral blood and cultured HUVEC cells were extracted by Trizol (Invitrogen, California, USA) method. Then, RNA was reverse transcribed into cDNA. For the detection of miR-16b, the primers for miR-16b were 5’- AGCAGCACGTAAATATTGG-3’ and universal primer. The 20 μl reaction system included 10 μl qRT-PCR-Mix, 0.5 μl forward primer, 0.5 μl reverse primer, 2 μl cDNA template and 7 μl ddH2O. The cycle conditions were the following: 95°C for 10 min, and followed by 40 cycles of 95°C for 1 min, 60°C for 30 s. The relative expression level was calculated by the 2- ΔΔCT method.
For detection of autophagy related 14 (AGT14) mRNA, the primers for ATG14 were as follows: Forward: 5- TTCATGCCGCTCTGTCGTAG-3 and Reverse: 5- GGATCTGCGCTGCATTTCTG-3. The reaction system included 10 μl qRT-PCR-Mix, 0.5 μl forward primer, 0.5 μl reverse primer, 5 μl cDNA template and 14 μl ddH2O. The PCR procedures were as follows: 95°C for 10 min, 40 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 20 s, and a final extension at 72°C for 2 min. The relative expression level was calculated by the 2-ΔΔCT method.
Transfection of miR-16b inhibitor to HUVEC cells
The HUVEC cells were divided into Control group, Negative Control (NC) group, and miR-16b inhibitor group. The cells were cultured in antibiotics free DMEM medium containing 10% FBS. When cell confluence reached about 70%-90%, the cells were used for transfection. Totally 1.25 ul of 25 pmol/ul miR-16 inhibitor and 1 ul of Lipo 2000 (Invitrogen Co., Carlsbad, CA, USA) were added into EP tubes containing 50 ul DMEM medium, respectively. The 2 tubes were mixed together after 5 min incubation at room temperature. The mixture was added to each well after incubation for 20 min at room temperature. After 6 h, fresh DMEM with 10% FBS was used to replace medium, then cultured for 72 h.
CCK-8 assay
After transfection with miR-16b, each group of cells were washed by PBS twice and digested by trypsin. The cells were then seeded in 96-well plate. Each well contained about 2 × 103 cells and each well had 3 replicates. At 24 h, 48 h and 72 h, CCK-8 solution was added into each well and incubated for 1 h at 37°C. The absorbance of cells was measured at 450 nm wavelength to estimate the proliferation of cells.
Flow cytometry analysis of apoptosis
Each group of HUVEC cells was collected by centrifugation after trypsin digestion, and then washed by pre-cooled PBS twice. The apoptosis was detected with the ANXN V FITC APOPTOSIS DTEC KIT I (BD Biosciences, Franklin Lakes, NJ, USA) based on the standard protocol of manufactures. Briefly, cells were collected by trypsin digestion. After centrifugation at 500 g for 5 min, the cell pellet was collected. After washing with pre-cooled PBS for 3 times, cells were incubated with Annexin V and PI in the dark. Cell apoptosis was detected by flow cytometry.
Flow cytometry analysis of cell cycle
After transfection for 48 h, cells of each group were digested by trypsin and washed by pre-cooled PBS twice. Then, 1 × 106 cells were used for cell cycle analysis based on standard protocol of BD cell Cycle/DNA Kits (BD Biosciences, Franklin Lakes, NJ, USA) and Templates kit (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, cells were collected and washed with pre-cooled PBS. Then, cells were incubated with 200 μl of Solution A at room temperature. After incubation for 10 min, 120 μl of Solution B was added and incubated at room temperature for 15 min. After washing, cells were incubated with 10 μl of Solution C in the dark at room temperature for 10 min. Finally, cell cycle was analysed by flow cytometry. Modfit software (BD Biosciences, Franklin Lakes, NJ, USA) was used to analyse cell cycle changes.
Bioinformatics prediction and dual luciferase validation
Target Scan software was used to analyse target genes of miR-16. For validating whether miR-16 can bind to ATG14, dual luciferase assay was applied. Based on the bioinformatics prediction, the wild-type 3’ UTR and the mutant 3’ UTR of ATG14 were synthesized in vitro and were cloned into the downstream of pMIR-REPORT luciferase vector by Spe-1 and Hind III enzyme. HEK293T cells were co-transfected with agomiR-16 (100 nM) mimics and wild-type ATG14 3’ UTR or the mutant 3’ UTR. After transfection for 24 h, cells were lysed and luciferase intensity was measured by GloMax 20/20 luminometer (Promega, Madison, WI, USA) based on the standard protocol of the luciferase kit (Promega, Madison, WI, USA). The intensity of Renilla was used as control, and the fluorescence intensity in different groups was analysed.
Western blot
After transfection for 48 h, the HUVEC cells were washed by pre-cooled PBS twice, and then RIPA lysis buffer containing 1% PMSF was added for lysis for 5 min on ice. Then, protein was loaded into 10% SDS-PAGE (constant voltage 100 V) and transferred to PVDF membrane. After blocking by 5% non-fat milk, the primary antibodies of AGT14 (1:800; rabbit antihuman polyclonal antibody; Cat# ab139727), Beclin1 (1:800; rabbit anti-human polyclonal antibody; Cat# ab55878), microtubule-associated protein 1 Light Chain 3 Beta (LC3B) (1:800; rabbit anti-human polyclonal antibody; Cat# ab48394) and GAPDH (1:5000; mouse anti-human monoclonal antibody; Cat# ab8245) were added and incubated at 4°C overnight. The secondary antibodies of HRP-conjugated goat anti-mouse (1:8000; Cat# ab6789) and goat anti-rabbit IgG (1:8000; Cat# ab6721) were added incubated at room temperature for 1 h. All the antibodies were purchased from Abcam Company (Boston, USA). Finally, the membrane was developed by enhanced chemiluminescence reagent. The developed film was scanned using the Alpha Imager gel imaging systems (Alpha Imager, Santa Clara, California, USA). And the Western blot images were analysed using Quantity One software (Bio-Rad Laboratories, Hercules, CA, USA). GAPDH was used as an internal control.
Analysis of autophagy by electron microscope
After transfection with miR-16 inhibitor for 48 h, cells were collected and were fixed by 2.5% glutaraldehyde at 4°C overnight. After washing once with PBS, the 1% osmium tetroxide was added and incubated for 1 h. Then electron microscopy was used to observe autophagy bodies.
Statistical analysis
The SPSS 16.0 software was used to do statistical analysis. All the data were shown as the mean ± SD. Standard t-test was used to compare difference between groups. P<0.05 was considered as statistically significant.
Results
MiR-16 is up-regulated in peripheral blood of patients with CHD
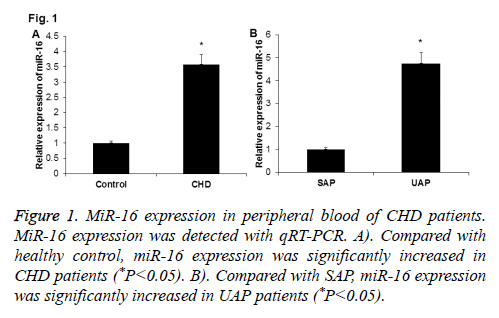
To investigate the expression of miR-16 in peripheral blood of CHD patients, qRT-PCR was performed. As shown in Figure 1A, miR-16 expression was significantly up-regulated in peripheral blood of CHD patients when compared with normal control (P<0.05). And, miR-16 expression was significantly higher in peripheral blood of UAP patients than in SAP patients (P<0.05) (Figure 1B). These results indicate that miR-16 may play roles in the development of CHD.
Figure 1: MiR-16 expression in peripheral blood of CHD patients. MiR-16 expression was detected with qRT-PCR. A). Compared with healthy control, miR-16 expression was significantly increased in CHD patients (*P<0.05). B). Compared with SAP, miR-16 expression was significantly increased in UAP patients (*P<0.05).
Effects of miR-16 on proliferation of HUVEC
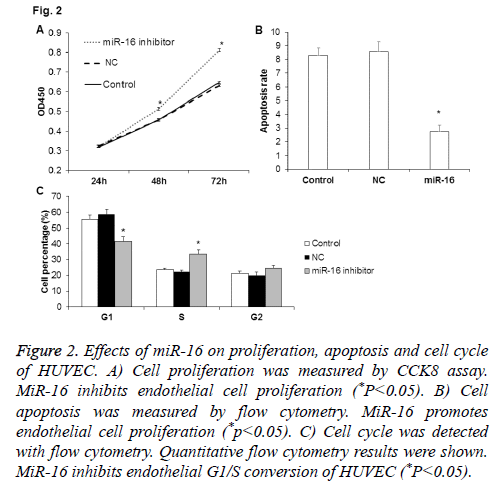
To check the influence of miR-16 on HUVEC proliferation, miR-16 expression was inhibited in HUVEC after transfection with miR-16 inhibitor and then CCK-8 assay was used to detect the changes of cell proliferation. As shown in Figure 2A, compared with NC and control, cell proliferation of HUVEC at 48 h and 72 h was significantly higher in cells transfected with miR-16 inhibitor (P<0.05). Thus, miR-16 may slow down the proliferation of HUVEC.
Figure 2: Effects of miR-16 on proliferation, apoptosis and cell cycle of HUVEC. A) Cell proliferation was measured by CCK8 assay. MiR-16 inhibits endothelial cell proliferation (*P<0.05). B) Cell apoptosis was measured by flow cytometry. MiR-16 promotes endothelial cell proliferation (*p<0.05). C) Cell cycle was detected with flow cytometry. Quantitative flow cytometry results were shown. MiR-16 inhibits endothelial G1/S conversion of HUVEC (*P<0.05).
Effects of miR-16 on apoptosis of HUVEC
To check the influence of miR-16 on HUVEC apoptosis, we inhibited miR-16 expression in HUVEC after transfection with miR-16 inhibitor. Then PI/ANNEXIN double staining method was used to detect the changes of cell apoptosis. As shown in Figure 2B, the apoptosis rate in miR-16 inhibitor group was significantly lower than NC group and control group (P<0.05), which indicate that miR-16 may promote the apoptosis of HUVEC.
Effects of miR-16 on cell cycle of HUVEC
To determine the effect of miR-16 on HUVEC cell cycle, flow cytometry analysis was performed. As shown in Figure 2C, the percentage of cells in G1 phase was significantly lower and that in S phase was significantly higher in miR-16 inhibitor group (P<0.05). The results indicate that miR-16 may inhibit proliferation of HUVEC through inhibiting G1/S transition.
ATG14 is directly targeted by miR-16
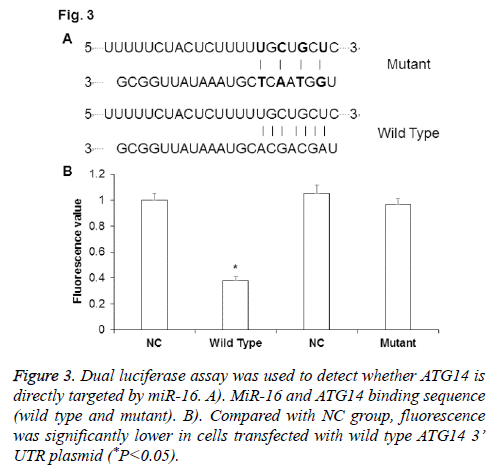
To determine whether ATG14 was directly targeted by miR-16, dual luciferase assay was performed after co-transfection of agomiR-16 and pMIR-REPORT plasmid. The binding sequence of miR-16 on ATG14 was shown in Figure 3A. As shown in Figure 3B, fluorescence value was significantly down-regulated in cells co-transfected with agomiR-16 and pMIR-REPORT-ATG14-wild type plasmid than in negative control group (P<0.05), while there was no significant difference between group co-transfected with agomiR-16 and pMIR-REPORT-ATG14-mutant and negative control group (P>0.05). The results from dual luciferase assay indicate that miR-16 can regulated ATG14 expression through complementary binding to 3’-UTR of ATG14 mRNA.
ATG14 mRNA level is regulated by miR-16
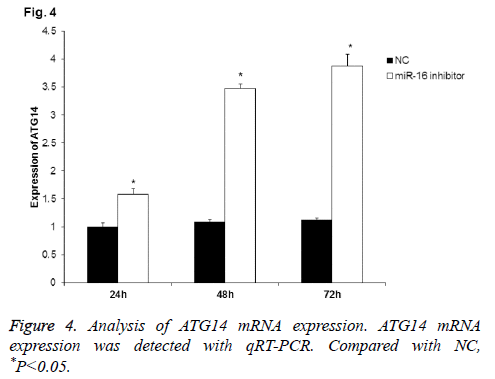
To further detect the correlation of miR-16 with ATG14, qRT-PCR was applied to detect ATG14 mRNA expression after miR-16 inhibition. As shown in Figure 4, ATG14 mRNA expression in HUVEC was gradually increased at 24 h, 48 h, and 72 h after transfection with miR-16 inhibitor. Statistically, there was significant difference between NC group and miR-16 inhibitor group at all-time points (P<0.05). The results further validated that miR-16 negatively regulated ATG14 expression in HUVEC.
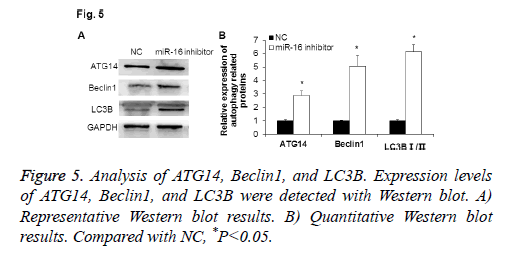
Expression of ATG14 and autophagy associated proteins by Western blot
To explore the function of miR-16 in HUVEC, Western blot was used to detect the expressions of autophagy related proteins after abnormally expressing miR-16. As shown in Figure 5, ATG14 protein expression was significantly increased when down-regulating miR-16 (P<0.05). The autophagy associated proteins Beclin1 was up-regulated, and the ratio of LC3B I/II was significantly increased (P<0.05). These results indicate that miR-16 may suppress cell autophagy of HUVEC.
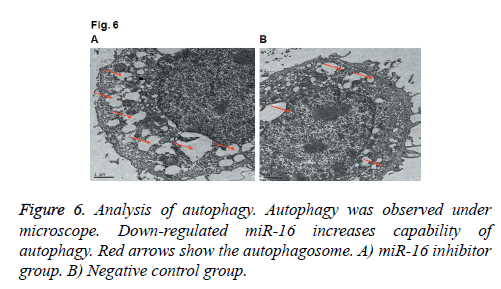
Effects of miR-16 on the formation of autophagosome by electron microscopeWe also applied electron microscope to observe the autophagosome in HUVEC. As shown in Figure 6, we found the number of autophagosomes was increased after down-regulating miR-16, which indicated that autophagy level was increased. The results indicate that miR-16 may affect autophagy of HUVEC.
Discussion
Vascular endothelial cell damage is an initiation process of atherosclerosis, which further leads to CHD, stroke and other complications [20]. It is reported that a variety of miRNA are changed during processes of cardiovascular diseases. For example, Wang et al. found that the expression levels of miR-1, miR-133a, miR-499, and miR-208a were lower in plasma of healthy human, while these expression levels were significantly increased in peripheral blood of acute myocardial infarction patients [21]. Among these miRNAs, miR-208a has better sensitivity and specificity in the diagnosis of myocardial infarction [21]. Other study demonstrated that miR-122 can be used as an early diagnosis marker for liver injury as it was significantly increased in plasma in patients with liver tissue damage [22]. In this study, the expression and the role of miR-16 expression in patients with CHD were analysed.
The progression of CHD is accompanied with aggravation of vascular endothelial cell damage [23]. Studies show that miRNA plays an important role in vascular endothelial cell damage. For example, Hu et al. reported that miR-21 was closely related with the proliferation of vascular endothelial cells [24]. Xu et al. found that miR-103 affected the viability of HUVEC cells through regulating Bcl-2 [25]. Importantly, it is shown that miR-16 could inhibit the cell proliferation of nasopharyngeal carcinoma cells and human cervical cancer cells [26,27]. However, the role of miR-16 in CHD was unknown. Our results showed that miR-16 expression was significantly increased in peripheral blood of patients with CHD than healthy people. And, miR-16 expression was significantly higher in peripheral blood of UAP patients than SAP patients, which indicated that miR-16 might be involved in the progression of CHD. We further studied the biological functions of miR-16 in endothelial cells. After down-regulating miR-16, HUVEC proliferation was promoted and G1/S conversion was also accelerated. Consistent with previous studies [26,27], the results indicated that the proliferation capability of endothelial cells was inhibited by miR-16, which might slow down the damage repair of endothelial cell injury.
Zhang et al. found that miR-16 inhibited the apoptosis of PC12 cells [28]. MiR-16 can regulate chemo resistance of human glioma cells by regulating Bcl-2 [29]. In this study, apoptosis analysis demonstrated that miR-16 promoted the apoptosis of endothelial cells. Together, we speculated that the highly expressed miR-16 strengthened endothelial cell damage.
Furthermore, we found that ATG14 was one of target genes of miR-16 by bioinformatics analysis. ATG14 is one of the autophagy related genes, which plays important roles in the development of autophagy [30]. We hypothesized that miR-16 might inhibit cell proliferation, promote apoptosis and inhibit cell autophagy. After transfection with miR-16 inhibitor, we found that autophagy associated genes (Beclin1, LC3B, and ATG14) were up-regulated, which indicated that cell autophagy was increased. The electron microscopy observation showed that the number of autophagosomes was significantly increased after down-regulating miR-16, which further validated that miR-16 can inhibit the development and progression of cell autophagy.
It is of great importance to identify molecular markers for diagnosis of CHD. For example, soluble ST2, a receptor of IL-33 may be used as a novel cardiac bio-marker [31,32]. Whether miR-16 could be used as a diagnostic marker for CHD is unknown and needs further investigation.
In summary, up-regulated miR-16 in peripheral blood was related to the development and progression of CHD.
Acknowledgements
The authors want to thank Dr. Bing Xu (Department of Emergency, Shanghai Ninth People's Hospital, School of Medicine, Shanghai Jiao Tong University) for his valuable help.
Disclosures
All authors declare no financial competing interests.
All authors declare no non-financial competing interests.
References
- Ladapo JA, Goldfeld KS, Douglas PS. Projected morbidity and mortality from missed diagnoses of coronary artery disease in the United States. Int J Cardiol 2015; 195: 250-252.
- Malik VS, Chiuve SE, Campos H, Rimm EB, Mozaffarian D, Hu FB, Sun Q. Circulating very-long chain saturated fatty acids and incident coronary heart disease in US men and women. Circulation 2015; 132: 260-268.
- Patel J, Al Rifai M, Blaha MJ, Budoff MJ, Post WS, Polak JF, Bluemke DA, Scheuner MT, Kronmal RA, Blumenthal RS, Nasir K, McEvoy JW. Coronary artery calcium improves risk assessment in adults with a family history of premature coronary heart disease: results from multi-ethnic study of atherosclerosis. Circ Cardiovasc Imaging 2015; 8: e003186.
- Liang Y, Li YP, He F, Liu XQ, Zhang JY. Long-term, regular remote ischemic preconditioning improves endothelial function in patients with coronary heart disease. Braz J Med Biol Res 2015; 48: 568-576.
- Cung H, Aragon MJ, Zychowski K, Anderson JR, Nawarskas J, Roldan C, Sood A, Qualls C, Campen MJ. Characterization of a novel endothelial biosensor assay reveals increased cumulative serum inflammatory potential in stabilized coronary artery disease patients. J Transl Med 2015; 13: 99.
- Domigan CK, Warren CM, Antanesian V, Happel K, Ziyad S, Lee S, Krall A, Duan L, Torres-Collado AX, Castellani LW, Elashoff D, Christofk HR, van der Bliek AM, Potente M, Iruela-Arispe ML. Autocrine VEGF maintains endothelial survival through regulation of metabolism and autophagy. J Cell Sci 2015; 128: 2236-2248.
- Barr MP, Gray SG, Gately K, Hams E, Fallon PG, Davies AM, Richard DJ, Pidgeon GP, O Byrne KJ. Vascular endothelial growth factor is an autocrine growth factor, signalling through neuropilin-1 in non-small cell lung cancer. Mol Cancer 2015; 14: 45.
- Dmitrieva NI, Burg MB. Elevated sodium and dehydration stimulate inflammatory signaling in endothelial cells and promote atherosclerosis. PLoS One 2015; 10: e0128870.
- Zhou JH, Pan ZY, Zhang YF, Cui WY, Long CL, Wang H. Stimulation of endothelial non-neuronal muscarinic receptor attenuates the progression of atherosclerosis via inhibiting endothelial cells activation. Zhongguo Ying Yong Sheng Li XueZaZhi 2014; 30: 549-559.
- Tian J, Hu S, Wang F, Yang X, Li Y. PPARG, AGTR1, CXCL16 and LGALS2 polymorphisms are correlated with the risk for coronary heart disease. Int J ClinExpPathol 2015; 8: 3138-3143.
- Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/ß-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur J Cancer 2015; 51: 1638-1649.
- Lukasik A, Wojcikowski M, Zielenkiewicz P. Tools4miRs - one place to gather all the tools for miRNA analysis. Bioinform 2016; 32: 2722-2724.
- Staszel T, Zapal AB, Polus A, Sadakierska-Chudy A, KieaWilk B. Role of microRNAs in endothelial cell pathophysiology. Pol Arch Med Wewn 2011; 121: 361-366.
- Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? ArteriosclerThrombVascBiol 2011; 31: 2383-2390.
- Caporali A, Emanueli C. MicroRNA-503 and the extended microRNA-16 family in angiogenesis. Trends Cardiovasc Med 2011; 21: 162-166.
- Gu Y, Wang XD, Lu JJ, Lei YY, Zou JY. Effect of mir-16 on proliferation and apoptosis in human A549 lung adenocarcinoma cells. Int J ClinExp Med 2015; 8: 3227-3233.
- Dwivedi SK, Mustafi SB, Mangala LS, Jiang D, Pradeep S, Rodriguez-Aguayo C, Ling H, Ivan C, Mukherjee P, Calin GA, Lopez-Berestein G, Sood AK, Bhattacharya R. Therapeutic evaluation of microRNA-15a and microRNA-16 in ovarian cancer. Oncotarget 2016; 7: 15093-15104.
- Chamorro-Jorganes A, Araldi E, Penalva LO, Sandhu D, Fernandez-Hernando C, Suarez Y. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. ArteriosclerThrombVasc Biol 2011; 31: 2595-2606.
- Huang S, Zou X, Zhu JN, Fu YH, Lin QX, Liang YY, Deng CY, Kuang SJ, Zhang MZ, Liao YL, Zheng XL, Yu XY, Shan ZX. Attenuation of microRNA-16 de-represses the cyclins D1, D2 and E1 to provoke cardiomyocyte hypertrophy. J Cell Mol Med 2015; 19: 608-619.
- Tison GH, Guo M, Blaha MJ, McClelland RL, Allison MA, Szklo M, Wong ND, Blumenthal RS, Budoff MJ, Nasir K. Multisite extra-coronary calcification indicates increased risk of coronary heart disease and all-cause mortality: The multi-ethnic study of atherosclerosis. J CardiovascComputTomogr 2015; 9: 406-414.
- Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microrna: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010; 31: 659-666.
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 2009; 106: 4402-4407.
- Watt J, Kennedy S, Ahmed N, Hayhurst J, McClure JD, Berry C, Wadsworth RM, Oldroyd KG. The relationship between oxidised LDL, endothelial progenitor cells and coronary endothelial function in patients with CHD. Open Heart 2016; 3: e000342.
- Hu J, Ni S, Cao Y, Zhang T, Wu T. The Angiogenic Effect of microRNA-21 Targeting TIMP3 through the Regulation of MMP2 and MMP9. PLoS One 2016; 11: e0149537.
- Xu MC, Gao XF, Ruan C, Ge ZR, Lu JD. miR-103 regulates oxidative stress by targeting the BCL2/adenovirus E1B 19 kDa interacting protein 3 in HUVECs. Oxid Med Cell Longev 2015; 2015: 489647.
- He Q, Ren X, Chen J, Li Y, Tang X. miR-16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signalling pathways. Oncotarget 2016; 7: 3047-3058.
- Zubillaga-Guerrero MI, Alarcon-Romero Ldel C, Illades-Aguiar B, Flores-Alfaro E, Bermúdez-Morales VH, Deas J, Peralta-Zaragoza O. MicroRNA miR-16-1 regulates CCNE1 (cyclin E1) gene expression in human cervical cancer cells. Int J ClinExp Med 2015; 8: 15999-16006.
- Zhang B, Chen CF, Wang AH, Lin QF. MiR-16 regulates cell death in Alzheimers disease by targeting amyloid precursor protein. Eur Rev Med PharmacolSci 2015; 19: 4020-4027.
- Han J, Chen Q1. MiR-16 modulates temozolomide resistance by regulating BCL-2 in human glioma cells. Int J ClinExpPathol 2015; 8: 12698-12707.
- Xu D, Wang Z, Chen Y. Two-layer regulation of PAQR3 on ATG14-linked class III PtdIns3K activation upon glucose starvation. Autophagy 2016; 12: 1047-1048.
- Ciccone MM, Cortese F, Gesualdo M, Riccardi R, Di Nunzio D. A novel cardiac bio-marker: ST2: a review. Molecules 2013; 18: 15314-15328.
- Marzullo A, Ambrosi F, Inchingolo M, Manca F, Devito F. ST2L transmembrane receptor expression: an immunochemical study on endarterectomy samples. PLoS One 2016; 11: e0156315.