ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 5
Selection of reliable reference genes for qRT-PCR analysis on head and neck squamous cell carcinomas
1Department of Medical Genetics, Cerrahpasa Faculty of Medicine, İstanbul University, İstanbul, Turkey
2Department of Medical Genetics, Faculty of Medicine, Selcuk University, Konya, Turkey
3Department of Medical Biology, Meram Faculty of Medicine, Necmettin Erbakan University, Konya, Turkey
4Department of Biostatistics, Karabuk University, Faculty of Medicine, Karabük, Turkey
5Department of Otorhinolaryngology, Faculty of Medicine, Selcuk University, Konya, Turkey
6Department of Genetics, Faculty of Veterinary Medicine, Selcuk University, Konya, Turkey
- *Corresponding Author:
- Aysel Kalayci Yigin
Department of Medical Genetics
Cerrahpasa Faculty of Medicine
İstanbul University, Turkey
Accepted date: June 23, 2016
The choice of reliable reference genes as an internal control is inevitable to obtain accurate results. Here we present an assessment of 7 reference genes (18S rRNA, 28S rRNA, ACTB, GAPDH, TUBA1, YWHAZ, and SDHA) to normalize gene expression data in Head and Neck Squamous Cell Carcinomas (HNSCCs). We attempted to determine a reliable set of reference genes to use in the normalization of gene expression data in Head and Neck Squamous Cell Carcinomas (HNSCCs) and normal mucosal tissues. Malignant and non-malignant tissue samples were collected from 12 patients with primary untreated HNSCC. geNorm and NormFinder software packages were used for data evaluations. Results obtained by geNorm indicated that average expression stability values (M) of all candidates genes were smaller than 1.5 (accepted M value for geNorm), showing that all the evaluated genes can be employed as HKGs, although GAPDH and ACTB were reported to be the most stable. Similarly, NormFinder results were in agreement with geNorm’s results. GAPDH and ACTB are considered to be most suitable reference genes to evaluate novel gene expression in the tissues several of HNSCCs.
Keywords
Housekeeping gene, geNorm, NormFinder, Head and neck squamous cell carcinoma.
Introduction
Reverse transcriptase quantitative real-time polymerase chain reaction (RT-qPCR) is a variation of the PCR, which is comprehensible, rapid, and sensitive technique in molecular biology [1,2]. This technique has been widely used to detect quantitative and precise measurement of the gene expression at the mRNA level in cancer molecular biogenesis field. In order to analyse the gene expression data obtained from RT-qPCR, crucial variations of RT-qPCR parameters must be considered, including amount of starting material, primer design, and RNA quality [1]. When assessing the gene expression levels between different samples, such variations should be minimized. Therefore, the normalization of a gene of interest to a reference gene, also known as a Housekeeping Gene (HKG), should be carried out carefully [3]. In recent decades, a few experimental methods have been used to select the most appropriate reference gene for normalization. However, to avoid possible errors in gene expression studies, the normalization of RTqPCR data to a reference gene subjected to the same preparation procedures as the candidate genes is considered the most reliable method [1,4,5].
A reference gene should exhibit stable expression levels in both normal and tumor tissues, and its expression level should not differ by the physiological, pathological, or other external causes [5,6]. However, no certain universal reference gene has been found. Additionally, internal and external factors may modify housekeeping gene expression [1,7].
With the validation of a possible reference gene for each study, researchers have disregarded the significance of validation of reference genes in some of the qPCR experiments, leading to erroneous conclusions [8,9]. Therefore, determination and validation of genes as internal controls are crucial steps in ensuring the correct variability among the samples in RT-qPCR [10].
In this study, we attempted to determine a reliable set of reference genes to use in the normalization of gene expression data in Head and Neck Squamous Cell Carcinomas (HNSCCs) and normal mucosal tissues.
Materials and Methods
Patients and sample collection
Tumor and non-tumor tissue samples were collected from 12 patients with primary untreated larynx and tongue squamous cell carcinomas. A small piece of biopsy samples was taken during the operation and sent to the pathology laboratory. The samples were frozen immediately and kept in liquid nitrogen at -196°C. Non-malignant tissues were also obtained from the same anatomical site. Tissues were separated as from the primary tumor lesions, spanning the midline and on the opposite side of the well-lateralized tumors.
RNA isolation, quality control and cDNA synthesis
Forty mg of each tissue samples was chopped with a homogenizer (Heidolph Silent Crusher M, Germany) in TRIzol (Ambion, USA). Total RNA extraction was carried out as recommended by manufacturer’s protocol. Purity of RNA was confirmed by optic density of 260/280 (2.0 ± 0.1) with Colibri microvolume spectrometer (Titertek-Berthold-Germany). Integrity of RNA was verified through agarose gel electrophoresis. One μg of total RNA was treated with DNAse- I (Thermo Scientific, USA) to eliminate genomic DNA contamination. cDNAs were synthesized by both blend of oligodT and random hexamer primers in equal volume using a iScript Reverse Transcription Supermix (BioRad, USA) per manufacturer’s protocol.
Primers and real time PCR
Actin Beta (ACTB), 28S rRNA (28S), 18S rRNA (18S), succinate dehydrogenase complex subunit A (SDHA), tubulin alpha 1 (TUBA1), tyrosine 3-monooxygenase/tryptophan 5- monooxygenase activation protein (YWHAZ), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), primers were designed as previously published sequences (Table 1). Each real-time PCR run was established as follows: 7,5 SsoAdvanced™ Universal SYBR Green Supermix, (BioRad, USA), 1 μl cDNA, 5 pMol of each primers, and ddH2O up to 15 μl of final volume. Amplification conditions were comprised of an initial denaturation at 95°C for 8 min, followed by 42 cycles of denaturation, annealing, and amplification (94°C 15 s, 60°C 30 s, 72°C 45 s using a Light Cycler 480 II (Roche). Analysis of melting curve was carried out as follows: 95°C 1 min, and subsequently fluorescence detection was done at each 1°C increment between 54°C and 95°C. Each of run included a negative control as no cDNA template. To confirm the reaction specificity and amplification, real-time PCR products were run on 2,5 % agarose gel. From very beginning of RNA extraction to real-time PCR, the whole procedure was replicated twice as technical procedure. Malignant and non-malignant samples were always performed in the same run to circumvent inter-run variation. To ensure specificity of all RT-qPCRs, each run was verified through melting curve analysis of amplification products.
| Locus | Location | Primer sequence (5’ → 3’) | Amplicon size (bp) | Gene bank accession | Reference |
|---|---|---|---|---|---|
| 18S | 6p21.3 | ATGCGGCGGCGTTATTCC GCTATCAATCTGTCAATCCTGTCC |
204 | AJ311673 | 19 |
| 28S | 19p13.2 | CGGGTAAACGGCGGGAGTAAC TAGGTAGGGACAGTGGGAATCTCG |
109 | EU554425 | 19 |
| ACTB | 7p15-p12 | TGGCTGGGGTGTTGAAGGTCT AGCACGGCATCGTCACCAACT |
238 | NM_001101 | 18 |
| GAPDH | 12p13 | ATCACCATCTTCCAGGAGCGAGA GTCTTCTGGGTGGCAGTGATGG |
341 | NM_001163856 | 20 |
| TUBA1 | 12q13.12 | GCCCTACAACTCCATCCTGA ATGGCTTCATTGTCCACCA |
78 | AW260995 | 21 |
| YWHAZ | 8q23.1 | ATGCAACCAACACATCCTATC GCATTATTAGCGTGCTGTCTT |
178 | NM_001135702 | 22 |
| SDHA | 5p15 | AGCAAGCTCTATGGAGACCT TAATCGTACTCATCAATCCG |
200 | NM_004168 | 18 |
Table 1: List of primers used.
A novel statistical approach for selection of HKG
Number of methods has been suggested and several software solutions have been released in order to select the best HKGs. Of those, the software NormFinder (http://www.mdl.dk/publicationsnormfinder.htm) by Andersen et al., geNorm (http://medgen.ugent.be/jvdesomp/genorm) by Vandesompele et al. have been most widely used [11,12]. Vandesompele et al. employed variation (i.e. the standard deviation of pairwise log2-transformed expression ratios) and introduced the genestability measurement Mj for control gene j as the arithmetic mean of all pairwise variations as the gene-stability measurement [12]. However, Andersen et al. evaluated the systematic variation across the sample subgroups apart from overall expression variation [11]. In this study, we performed descriptive and univariate statistical analysis for candidate HKGs. We have used software packages geNorm and NormFinder in the selection of HKGs and results were compared. All statistic methods were previously discussed in studies by Kayis et al. [13].
Results
The expression stability of the seven candidate controls was assessed to establish the least variable reference genes by using geNorm and NormFinder software analyses. Descriptive statistics and Coefficient of Variation (CV (%)) are shown in Tables 2 and 3, respectively.
| Genes | Groups | Min. | Max. | SD | ||||
| TT | TN | LT | LN | OVERALL | SD | SD | Range | |
| 18S | 28.71 (± 0.41) | 28.58 (± 1.20) | 28.33 (± 0.82) | 28.92 (± 0.49) | 27.59 (± 0.55) | 0.41 | 1.21 | 0.8 |
| 28S | 27.94 (± 0.55) | 27.86 (± 0.74) | 25.08 (± 5.33) | 27.60 (± 0.46) | 28.92 (± 0.49) | 0.46 | 5.33 | 4.87 |
| ACTB | 30.11 (± 0.22) | 30.04 (± 0.12) | 29.65 (± 0.83) | 29.89 (± 0.59) | 29.92 (± 0.52) | 0.12 | 0.83 | 0.71 |
| GAPDH | 27.94 (± 0.39) | 27.68 (± 0.46) | 27.09 (± 0.50) | 27.67 (± 0.56) | 27.59 (± 0.55) | 0.39 | 0.56 | 0.17 |
| TUBA1 | 33.14 (± 1.34) | 31.52 (± 0.67) | 31.88 (± 0.79) | 31.20 (± 0.94) | 31.94 (± 1.17) | 0.67 | 1.34 | 0.67 |
| YWHAZ | 33.43 (± 1.26) | 31.11 (± 6.21) | 31.99 (± 1.91) | 33.13 (± 1.00) | 32.41 (± 3.26) | 1 | 6.21 | 5.21 |
| SDHA | 29.06 (± 1.21) | 29.33 (± 1.02) | 28.69 (± 1.13) | 29.13 (± 1.13) | 29.06 (± 1.07) | 1.02 | 1.21 | 0.19 |
Table 2: Mean and standard deviations (± SD) of Ct values of genes in between groups and overall.
| Genes | Groups | ||||
|---|---|---|---|---|---|
| TT | TN | LT | LN | Overall | |
| 18S | 1.43 | 4.21 | 2.91 | 1.72 | 2.71 |
| 28S | 2 | 2.66 | 21.24 | 1.69 | 10.34 |
| ACTB | 0.76 | 0.41 | 2.83 | 1.97 | 1.75 |
| GAPDH | 1.43 | 1.68 | 1.85 | 2.05 | 2.01 |
| TUBA1 | 4.05 | 2.15 | 2.48 | 3.03 | 3.69 |
| YWHAZ | 3.78 | 19.97 | 5.98 | 3.04 | 10.06 |
| SDHA | 4.18 | 3.49 | 3.95 | 3.91 | 3.71 |
Table 3: Coefficient of Variation (CV (%)) of Ct values of genes in between groups and overall.
All the HKGs had steady expression levels according to the geNorm software. The most steady genes were ACTB (M=0.028), and GAPDH (M=0.028) among seven HKGs, whereas the least steady genes were 28S (M=0.125) and YWHAZ (M=0.095). GAPDH had the lowest mean of Ct value (26.21), while YWHAZ had the highest (35.0).
The NormFinder software detected GAPDH (stability value=0.176) and ACTB (stability value=0.184) as the best and second best stably expressed genes, respectively, while YWHAZ (stability value=0.908) was found to be the least stable gene. GAPDH and ACTB were the best combination of genes as HKG, with a stability value of 0.127.
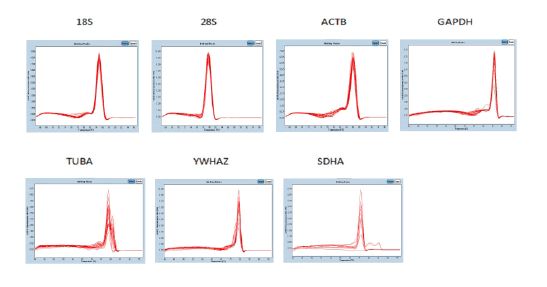
RT-qPCR specificity of all reactions was verified through melting curve analysis of the RT-qPCR products (Figure 1).
Discussion
Expression steadiness of seven candidate HKGs (Table 2) were established using different statistical procedures. Descriptive statistics have shown that the smallest values for maximum SD, SD range, and overall SD were obtained from GAPDH while the second smallest values for those three descriptive statistics were obtained from ACTB. The highest values for maximum SD, SD range, and overall SD were obtained from YWHAZ, and the second highest overall SD was observed in 28S. The lowest CV was obtained from ACTB, while the second lowest CV value was obtained from GAPDH. 28S and YWHAZ had the highest CV.
The results were also analysed with commonly used software packages geNorm and NormFinder. Out of all HKGs, the expression stability values (M) calculated via geNorm was less than 1.5, suggesting that these candidate genes had steady expression levels. Of seven HKGs, the most steady genes were ACTB (M=0.488), GAPDH (M=0.488), whereas the least stable gene was YWHAZ (M=0.886). The results of geNorm and NormFinder were consistent with each other.
The development of quantitative PCR and RT-PCR techniques has benefits for the incorporation of molecular methods into clinical practice. RT-qPCR is a very precise method for the evaluation of low abundance mRNAs and may be used for various applications [14]. For instance, these applications are comprehensive for molecular studies and include procedures designed to provide a molecular appreciation for staging malignancies [15], analysis of cytokine mRNA levels [16], identifying circulating tumor cells in cancer patients, and so on [17].
The data reliability of the quantitative RT-qPCR is important for molecular studies. Finding the total amount of present cDNA may be difficult in different samples, and results of the analysed genes are often normalized to a reference gene presumed to be invariant. This approach may not always be accurate. Since many of the genes are regulated in several conditions, there is no unique universal reference gene, which this may cause to altered findings and incorrect experimental outcomes. There should be a regular expression in different tested tissues for a reliable reference gene. Also, a reliable reference gene should not be regulated by physiologic or external causes.
The current study is the first report of a systematic evaluation of suitable reference genes for the normalization of RT-qPCR experiments in malignant tongue and larynx cancer expression studies. Although there has been a similar study related to the HNSCC [18], our study is comparatively notable, because the selected genes are variable and selected tissues are specific to the larynx and tongue.
Amongst seven commonly used classical housekeeping genes, the most stable genes were ACTB (M=0.028), and GAPDH (M=0.028), whereas the least stable gene was 28S (M= 0.125). The GAPDH had the lowest mean Ct value (26.21) whilst the YWHAZ had the highest (35.0) on the geNorm software program. Additionally, NormFinder detected GAPDH (stability value=0.176) and ACTB (stability value=0.184) as the best and second stably expressed genes, respectively, while the YWHAZ (stability value=0.908) was found to be the least stable gene. The very best set of two genes was GAPDH and ACTB, and their stability value was 0.127. The results of the both the software programs were highly comparable.
In conclusion, this study used a combination of the two methods (geNorm and NormFinder). It suggests that GAPDH and ACTB should be considered suitable reference genes when studying differences in gene expression profiles between tumour and non-tumour tissue samples in HNSCC.
References
- Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 2002; 30: 503-512.
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 2002; 29: 23-39.
- Karge WH, Schaefer EJ, Ordovas JM. Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol Biol 1998; 110: 43-61.
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 2005; 6: 279-284.
- Nygard AB, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol 2007; 8: 67.
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B. Housekeeping genes as internal standards: use and limits. J Biotechnol 1999; 75: 291-295.
- Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer 2008; 8: 20.
- Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GA, Zumla A. The implications of using an in appropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 2005; 344: 141-143.
- Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, Garrick T, Sheedy D, Hurd Y, Bakalkin G, Ekstrom TJ. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronical coholics. Brain Res 2007; 1132: 20-28.
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004; 37: 112-114, 116, 118-119.
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004; 64: 5245-5250.
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: 30034.
- Kayis SA, Atli MO, Kurar E, Bozkaya F, Semacan A, Aslan S, Guzeloglu A. Rating of putative housekeeping genes for quantitative gene expression analysis in cyclic and early pregnant equine endometrium. Anim Reprod Sci 2011; 1: 124-132.
- Bustin S. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000; 25: 169-193.
- Bustin SA, Dorudi S. Molecular assessment of tumour stage and disease recurrence using PCR-based assays. Mol Med Today 1998; 4: 389-396.
- Forlenza M, Kaiser T, Savelkoul HF, Wiegertjes GF. The use of real-time quantitative PCR for the analysis of cytokine mRNA levels. Methods Mol Biol 2012; 820: 7-23.
- Ghossein RA, Rosai J. Polymerase chain reaction in the detection of micrometastases and circulating tumor cells. Cancer 1996; 78: 10-16.
- Lallemant B, Evrard A, Combescure C, Chapuis H, Chambon G, Ravnal C, Reynaud C, Sabra o, Joubert D, Hollande F, Lallemant JG, Lumbroso S, Brouillet JP. Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Mol Biol 2009; 10: 78.
- Zhang, YW, Davis EG, Bai J. Determination of internal control for gene expression studies in equine tissues and cell culture using quantitative RT-PCR. Vet Immunol Immunopathol 2009; 130: 114-119.
- Boerboom D, Brown KA, Vaillancount D, Poitras P, Goff AK, Watanabe K, Dore M, Sirois J. Expression of key prostaglandin synthases in equine endometrium during late diestrus and early pregnancy. Biol Reprod 2004; 70: 391-399.
- Bogaert L, Van Poucke M, De Baere C, Peelman L, Gasthuys F. Selection of a set of reliable reference genes for quantitative real-time PCR in normal equine skin and in equine sarcoids. BMC Biotechnol 2006; 6: 24.
- Erkens T, Poucke MV, Vandesompele J, Goossens K, Zeveren AV, Peelman LJ. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotech 2006; 6: 41.