ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2009) Volume 20, Issue 2
Superoxide dismutase : An alternate target for Plasmodium
1Department of Biochemistry, Yenepoya Medical College, Yenepoya University, Deralakatte, Mangalore 575018
2Department of Microbiology, Yenepoya Medical College, Yenepoya University, Deralakatte, Mangalore 575018
3Department of Biochemistry, Yenepoya Medical College, Yenepoya University, Deralakatte, Mangalore 575018 Karnataka, India
- *Corresponding Author:
- Vinitha R. Pai
Department of Biochemistry
Yenepoya Medical College
Yenepoya University Deralakatte
Mangalore 575018, Karnataka, India
Phone: +91 9980145182
E- mail: vinitharpai(at)gmail.com
Accepted March 02 2009
Superoxide dismutases (SOD’s) are key enzymes which catalyze the dismutation of the O2- free radical to hydrogen peroxide and oxygen. Hence SOD is an important enzymatic, antioxidant defense in all cells exposed to oxygen. Two different SODs have been described in mammalian cells: cytosolic, Cu-Zn containing enzyme, and, mitochondrial Mn- dependant enzyme. A third Fe- containing SOD is known to occur in prokaryotes, protozoans, algae and higher plants. The FeSOD’s may play a role as antioxidant defense mechanisms in parasitic protozoans, such as Plasmodium falciparum (Pf), which are highly susceptible to superoxide oxidative stress. SOD may have a significant role in the life cycle of this parasite, especially in the erythrocytic phase, where reactive oxygen species can cause the degradation of hemoglobin. Therefore, an evaluation of the Fe-SODs as potential targets to antimalarial drugs was done by bio-informatic analysis. The protein sequences of Cu-ZnSOD1, MnSOD2 from H. sapiens and FeSOD of E. Coli were aligned with that of PfFeSOD in pairs or in three’s. The conserved amino acid residues were compared. The potential proteolytic sites were studied by using peptide cutter software to further confirm the similarity/dissimilarity in the primary structure of the two proteins. In conclusion, this study shows that the PfFeSOD is different from cytosolic Cu-ZnSOD1 of H. sapiens which is likely to be found in the erythrocytes, with just five conserved residues. There were few common proteolytic cleavage sites between the two primary sequences. Hence, the PfFeSOD could be an ideal enzyme to target anti-malarials for this parasite.
Keywords
Superoxide dismutase; Lactate dehydrogenase; Alignment; Multi-alignment;
Introduction
Superoxide dismutases (SOD’s) (Superoxide oxido-reductase: EC 1.15.1.1) have a wide distribution in nature and are indicated to be present in all oxygen metabolizing cells [1]. SOD’s are the key enzymes which catalyze the dismutation of the O2- free radical to hydrogen peroxide and oxygen.
2O2- + 2H+ → O2 + H2O2
It protects the oxygen metabolizing cells against the harmful effects of superoxide radicals and its derivatives such as peroxynitrite. Hence SOD is an important enzymatic, antioxidant defense in all cells exposed to oxygen Two different superoxide dismutases have been described in mammalian cells: a cytosolic, Cu-Zn containing enzyme SOD1, and, a Mn- dependant enzyme SOD2, located in mitochondria [3]. Other enzymes such as glutathione peroxidase and catalase present in all aerobic cells are also known to serve a protective function against oxidative damage. A third form of superoxide dismutase which contains iron is known to occur in prokaryotes, protozoans, algae and higher plants. This form of the enzyme is structurally different from the Cu-ZnSOD1 but has only a few differences with the MnSOD2 enzyme [4]. The FeSOD’s may play an important role as antioxidant defense mechanisms of many parasitic protozoans, such as Plasmodium falciparum, which are highly susceptible to superoxide oxidative stress.
Plasmodium falciparum, the protozoan parasite causing falciparum malaria is versatile when it comes to survival and defense strategies [5]. Two genes coding for FeSOD’s have been identified in the P. falciparum genome, which are very similar to the mitochondrial MnSOD of mammals. These forms of SOD’s may have a significant role in the life-cycle of the parasite, especially in the erythrocytic phase, where reactive oxygen species can cause the degradation of hemoglobin [6]. Therefore an evaluation of the FeSODs as potential targets to antimalarial drugs could be a new approach to chemotherapy of malaria. In the present study we have evaluated the differences between the SOD of H. sapiens and P. falciparum in comparison with lactate dehydrogenase (LDH). Though the lactate dehydrogenase enzymes of the compared species seems to be having a higher degree of similarity, the similarity of the PfFeSOD enzyme with that of Cu-ZnSOD of H. sapiens appears to be very negligible.
Materials and Methods
Bioinformatic approach was used to study the similarities and differences between the different SODs of this study.
The study was designed as follows
1. Protein sequences of Cu-ZnSOD1 [7], mitochondrial MnSOD2 [8], FeSOD of E. coli [9] and FeSOD of P. falciparum (PfFeSOD) [10] were obtained in FASTA format using the NCBI databank using the www.ncbi.nlm.nih.gov accession
2. The primary sequences were aligned in pairs using the ALIGN tool on the www.ebi.ac.uk/clustalw/ website, to study the percentage similarity/identity between the various forms to be evaluated. Identical residues are denoted by an asterix ’٭’ while similar residues are marked with a colon ’:’. When two amino
acid residues are the same when aligned against each other they are said to be ‘identical’. The residues are said to be similar when the aligned residues belong to the same class of amino acids. The percentage identity/ similarity was expressed as a total.
3. The conserved/invariant amino acid residues in the all the four sequences were studied by multialignment of all the sequences to be compared using www.ebi.ac.uk/clustalw/ website.
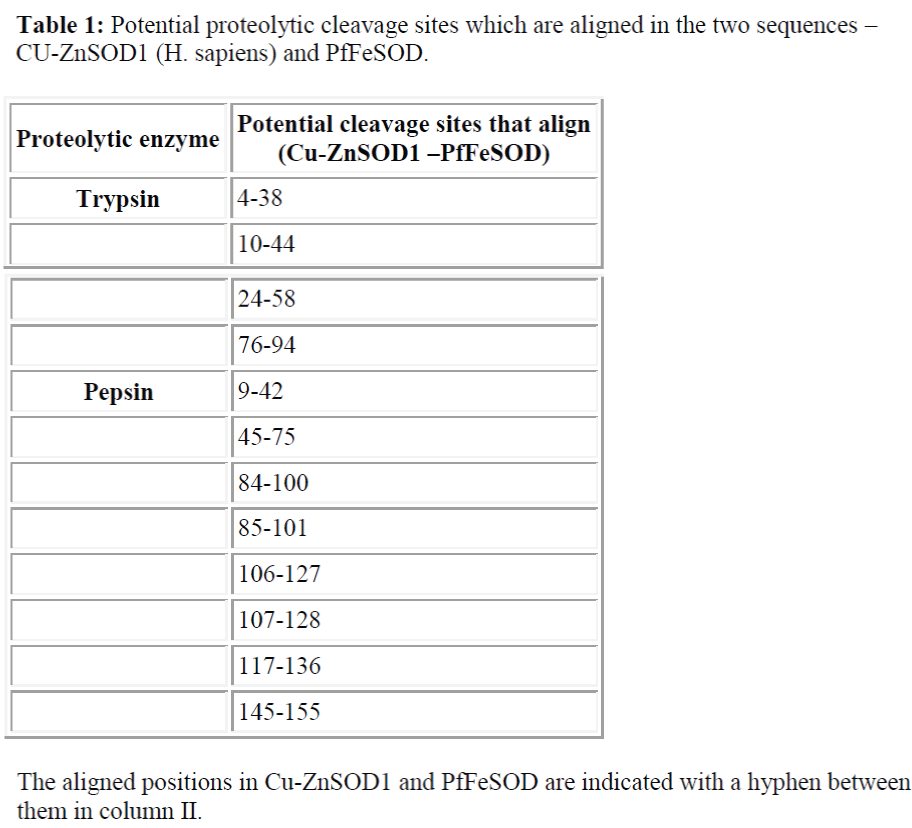
4. The potential cleavage sites of the proteolytic enzymes, pepsin and trypsin, in the Cu-ZnSOD1 and PfFeSOD were studied using the peptide cutter software. The common cleavage sites were listed in a table, sorted sequentially by amino acid numbers.
Results
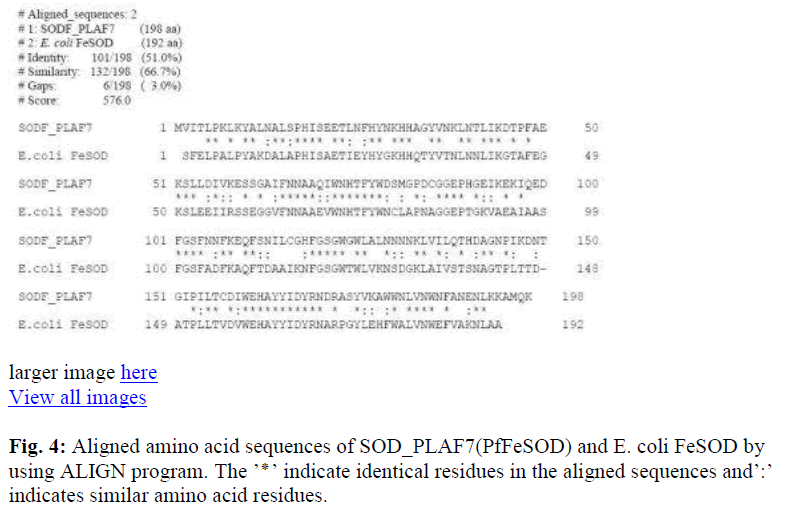
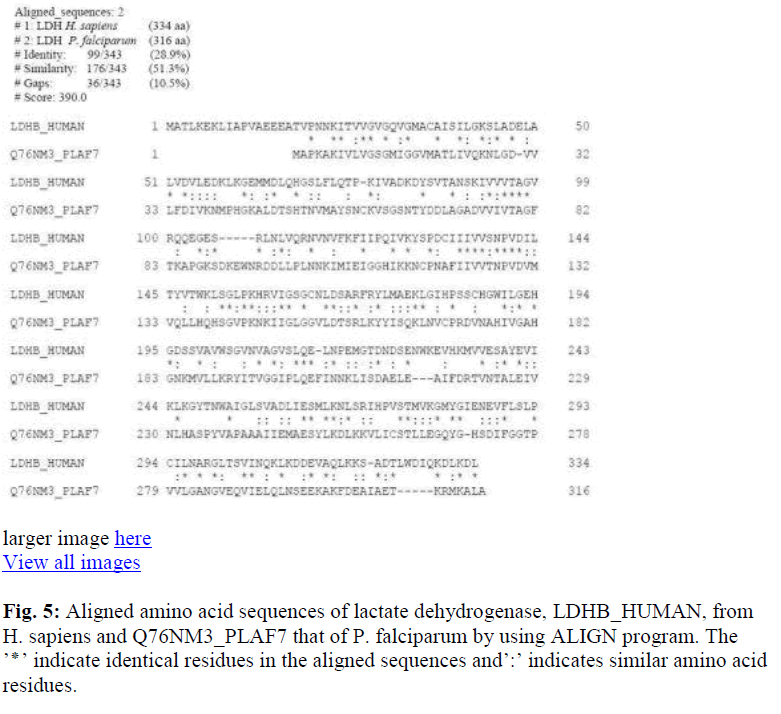
The primary sequences were first aligned in pairs to study the percentage identity / similarity of the compared sequences. The identity / similarity was expressed as total percentage since presence of similar amino acid residues (from the same group) in the aligned positions will not have much variation in the activity or higher structures of organization of the protein. Alignment of the mitochondrial MnSOD2 and cytosolic Cu-ZnSOD1 from H. sapiens (Fig. 1), showed identity / similarity of approximately 38%. PfFeSOD with the MnSOD of H. sapiens had a much higher degree of identity / similarity of about 83.7% (Fig. 2), Cu-ZnSOD1 of human with the PfFeSOD showed very little identity / similarity approximately 37% (Fig. 3). Alignment of the PfFeSOD with that of E. coli FeSOD, showed that the two are almost identical / similar (Fig. 4). Alignment of the lactate dehydrogenase enzymes of H. sapiens and PfFeSOD shows a very high sequence identity / similarity of about approximately 80% (Fig. 5). An alignment of three sequences – MnSOD2 of H. sapiens, E.coli FeSOD and PfFeSOD showed the presence of about 60 invariant / conserved amino acid residues distributed throughout the length of the three aligned sequences (Fig. 6). However, alignment of these three sequences with the Cu-ZnSOD1 of H. sapiens, the number of invariant residues is only 5 (Fig. 7).
The aligned positions in Cu-ZnSOD1 and PfFeSOD are indicated with a hyphen between them in column II.
Discussion
Plasmodium falciparum is a protozoan parasite causing falciparum malaria, which has varied survival and defence strategies. Multiple drug resistance is one of the strategies adopted by the asexual blood stages of this parasite [5]. Hence a search for alternate targets for therapy against this protozoan parasite will help us in tackling one of the dreaded diseases of mankind. The parasite developing resistance against the first and second line anti-malarial drugs has further necessitated the urgency of finding new drug targets and developing new pharmacophores to treat this dreaded disease [11].
The Fe- containing SOD which is present in the anaerobic bacteria and prokaryotes is considered to be the most primitive. Two other forms, Mn- containing form and Cu-Zn- containing forms are present in the mitochondria and cytosol of eukaryotic species [12] respectively. Structural studies indicate that FeSOD and MnSOD2 enzymes are similar in their primary, secondary and tertiary structures, while cytosolic Cu-ZnSOD1 form has no resemblance to these forms. The primary structures of FeSOD’s from E. coli [9], Psuedomonas ovalis [13], photobacterium leiogmathi [14] have been published. A comparison of the primary structures of six known MnSOD structures and three FeSOD has already been reported [4]. The FeSOD sequences of bacterial proteins showed a relatively high identity (65-74%). Saccharomyces cerevisiae stands apart from the compared sequences with a very low degree of identity [4].
The primary sequence of PfFeSOD is known. Hence it was of interest to see the degree of similarity between the MnSOD2 and Cu-ZnSOD1 of H. sapiens and FeSOD of E. coli bacteria with that of PfFeSOD for an understanding of the uniqueness of this enzyme which could probably make it an ideal target for anti-malarials. comparison of the two forms – mitochondrial MnSOD2 and cytosolic Cu-ZnSOD1 from H. sapiens is shown in fig. 1. The identity / similarity are approximately 38%.
While a comparison of the PfFeSOD with the MnSOD of H. sapiens had a much higher degree of identity / similarity of about 83.7% (Fig. 2). It was reported earlier that the Fe-SOD from other sources were structurally similar to that of MnSOD [4,15,16,17,18]. And in this study we found that PfFeSOD follows a similar pattern. However, since the asexual cycle of the plasmodium species is in the erythrocytes, which do not have the organelle mitochondria and the mitochondria associated MnSOD2 isoform, drug targeting of the PfFeSOD could probably be specific for the cause with no interfering effects, either from the mitochondrial MnSOD2 or from the cytosolic Cu-ZnSOD1.
Alignment of the Cu-ZnSOD1 of human (154 amino acid residues) with the PfFeSOD (198 amino acid residues) showed very little identity / similarity of approximately 37% (Fig. 3). Though the two are isozymes, they appear to be distinct proteins and the differences could be made use of in differentially targeting the parasite SOD from that of the host. The PfFeSOD had an N-terminal extension of 34 amino acids and C-terminal extension of 33 amino acids as compared to the Cu-ZnSOD1, while, the Cu-ZnSOD1 is aligned mostly to the central part of the primary structure of the PfFeSOD. Bernhardt/Pinto/Pai
Alignment of the PfFeSOD with that of E. coli bacteria, showed that the two are almost identical / similar (Fig. 4) while alignement with Cu-ZnSOD1 was only 37% (Fig. 3). This clearly showed that the PfFeSOD is a typical FeSOD of prokaryotes.
Now it was of interest to see whether primary sequences of other proteins from P. falciparum and H. sapiens also show this difference in identity / similarity or is this seen only in SOD. Alignment of the lactate dehydrogenase enzymes from these sources shows a very high sequence identity / similarity of about approximately 80% (Fig. 5). This observation clearly suggests that the sequence uniqueness seems to be restricted to the SOD enzyme of this protozoan parasite.
An alignment of the MnSOD2 of H. sapiens, E.coli FeSOD and PfFeSOD showed the presence of about 60 invariant / conserved amino acid residues distributed throughout the length of the three aligned sequences (Fig. 6). Tertiary structure of PfFeSOD is very similar to those of a number of Fe- and Mn- dependent SODs. Yet, comparison of the residues at the dimer interphase does show differences which can be played upon to design parasite specific drugs [6]. Most of these invariant residues have been speculated to be involved either in substrate recognition / binding or ligand binding or maintainance of aromatic nature of the active site or involved in subunit contact [4]. However, when these three sequences are aligned with the Cu-ZnSOD1 of H. sapiens, the number of invariant residues is only 5 clearly bringing out the differences between the cytosolic Cu-ZnSOD1 and PfFeSOD (Fig. 7). These five invariant residues do not have any major role like substrate binding, ligand binding or catalysis as compared to that of B. stearothermophilus [4]. Potential cleavage sites of protease enzymes trypsin and pepsin was done by using the peptide cutter. The results of this study showed that the PfFeSOD had 19 potential trypsin cleavage sites as compared to the 14 in Cu-ZnSOD1. Only 4 of these sites (21% PfFeSOD and 28% in Cu-ZnSOD1 of the total trypsin cleavage sites) are aligned against each other as shown in Table I. The potential pepsin cleavage sites was also 44 in PfFeSOD and 18 in Cu-ZnSOD1 out of which only 7 were aligned against each other (39% PfFeSOD and 16% in Cu-ZnSOD1 of the total pepsin cleavage sites) (Table I). Therefore the potential proteolytic cleavage sites predicted by the peptide cutter also show few similaritites between the Cu-ZnSOD1 of H. sapiens and PfFeSOD.
Further, the dendogram of the 4 compared sequences in this study (Fig. 8) confirms that the Cu-ZnSOD1 is most distant related to PfFeSOD.
In conclusion this study shows that the FeSOD in P. falciparum is a unique enzyme. It has very little similarity and identity with cytosolic Cu-ZnSOD1. The number of invariant / conserved amino acid residues in the CuZnSOD1 enzyme and the other Mn- and FeSODs compared in this study are only five. Common potential proteolytic enzyme cleavage sites in the two sequences is less. Since the parasite life-cyle has a phase in erythrocytes, the PfFeSOD could be a potential target for antimalarials, with no interference from the host erythrocytic Cu-ZnSOD1. Therefore specific inhibitors for the parasitic FeSOD could be developed for improved chemotherapy against malaria.
References
- Gregory EM, Stephen A Goscin SA, Fridovich I. Superoxide Dismutase and Oxygen Toxicity in a Eukaryote. Journal of Bacteriology 1974; 117: 2: 456-460.
- Fridovich I. Superoxide Radical and Superoxide Dismutases. Annual review of Biochemistry 1995; 64: 97-112.
- Richard A. Weisiger and Irwix Fridovich. Superoxide Dismutase – organelle specificity. The journal of Biological chemistry 1973; 248, 10: 3582-3592.
- Parker MW, Blake CC. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988; 229 (2): 377-82.
- Ramya T. N. C.; Surolia Namita and Avadesha Surolia. Survival strategies of the malarial parasite ‘Plasmodium falciparum’. Currrent Science 2002; 83: page numbers?
- Boucher IW, Brzozowski AM, Brannigan JA, Schnick C, Smith DJ, Kyes SA, Wilkinson AJ. The crystal structure of superoxide dismutase from Plasmodium falciparum. BMC Struct Biol. 2006; 4: 6-20.
- Kayatekin,C., Zitzewitz,J.A. and Matthews,C.R. Zinc binding modulates the entire folding free energy surface of human Cu,Zn superoxide dismutase. J. Mol. Biol. 2008; 384 (2): 540-555.
- Parker MW, Blake CC, Barra D, Bossa F, Schinina ME, Bannister WH, Bannister JV. Structural identity between the iron, and manganese-containing superoxide dismutases. Protein Eng. 1987; 1(5):393-7.
- Schininà ME, Maffey L, Barra D, Bossa F, Puget K, Michelson AM. The primary structure of iron superoxide dismutase from Escherichia coli. FEBS Lett. 1987;221(1):87-90.
- Baert CB, Deloron,P, Viscogliosi E, Dauchez M, Camus D, Dive D. Analysis of genetic diversity at the iron-containing superoxide dismutase locus in Plasmodium falciparum wild isolates. FEMS Microbiol. 1999; Lett. 181 (2): 237-243.
- Govindrajan Padmanabhan, Arun Nagaraj V and Pundi Rangarajan. Druga and drug targets against malaria. Current Science 2007; 92: page numbers?.
- McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968- 1988). Free Radic Biol Med. 1988;5 (5-6): 363-369.
- Isobe T, Fang YI, Muno D, Okuyama T, Ohmori D, Yamakura F. Amino acid sequence of iron-superoxide Superoxide dismutase: An alternate target for Plasmodium dismutase from Pseudomonas ovalis. FEBS Lett. 1987; 223(1):92-6.
- Barra D, Schininà ME, Bannister WH, Bannister JV, Bossa F. The primary structure of iron-superoxide dismutase from Photobacterium leiognathi. J Biol Chem. 1987; 262(3):1001-9.
- Ringe D, Petsko GA, Yamakura F, Suzuki K, Ohmori D. Structure of iron superoxide dismutase from Pseudomonas ovalis at 2.9-A resolution. Proc Natl Acad Sci U S A. 1983; 80(13):3879-83.
- Stallings WC, Pattridge KA, Strong RK, Ludwig ML. The structure of manganese superoxide dismutase from Thermus thermophilus HB8 at 2.4-A resolution. J Biol Chem. 1985 ; 260 (30): 16424-1632.
- Stallings WC, Pattridge KA, Strong RK, Ludwig ML. Manganese and iron superoxide dismutases are structural homologs. J Biol Chem. 1984; 259(17): 10695-1069.
- Stallings WC, Powers TB, Pattridge KA, Fee JA, Ludwig ML. Iron superoxide dismutase from Escherichia coli at 3.1-A resolution: a structure unlike that of copper/zinc protein at both monomer and dimer levels. Proc Natl Acad Sci U S A. 1983; 80(13) 3884-3888.