ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2019) Volume 30, Issue 1
Testing different concentrations of both hydrocortisone and rifampicin toward pregnane X receptor activation suggest their protective effect against liver toxicity.
Elham Alshammari*
Department of Pharmacy Practice, Princess Nourah bint Abdul Rahman University, Riyadh, Saudi Arabia
- *Corresponding Author:
- Elham Alshammari
Department of Pharmacy Practice
Princess Nourah bint Abdul Rahman University
Saudi Arabia
Accepted date: February 12, 2019
DOI: 10.35841/biomedicalresearch.30-19-069
Visit for more related articles at Biomedical ResearchIn different animal and human (in vitro) models, PXR activation resulted in suppressed liver inflammation and fibrosis which in return suggest graft survival. In this study, different concentrations of hydrocortisone (HC) and rifampicin were tested for PXR activity. For each drug, transfected HEPG2 cells were treated with different molar concentrations and then luciferase activity for both firefly and renilla were recorded. Both hydrocortisone and rifampicin produced an increase in PXR activity. Correlation between PXR activation and graft failure must be addressed in clinical setting
Keywords
PXR, Pregnane X receptor, Hydrocortisone, Rifampicin, Liver, Liver toxicity, Graft rejection
Introduction
Liver transplantation
Liver plays vital role in digestion, detoxification, blood clotting, storing of glucose, fat, iron and copper, and metabolism of medications and toxins [1]. Organ transplantation saves untreatable patients from end stage organ dysfunction. As a procedure, liver transplantation comes second after kidney transplantation. The procedure is easily described by replacing a diseased liver with a whole or part of healthy one from brain or heart dead donor (i.e. cadaveric) or using the right lobe from a live donor [2]. The worldwide statistics showed that 20, 200 liver transplants were performed since 2008 [3]. The most recent report published in Saudi Arabia showed that 930 liver transplants were performed since 1990 [4]. In addition to the response of the immune cells toward the new tissues, after liver transplantation ischemia and reperfusion injury are common and may put patients at risk of graft rejection [5]. In order to avoid graft rejection, patients receive number of medications like immunosuppressives. From one medical center to another, different protocols and treatment plans are used to maintain stability and survival after transplant [6].
Nuclear receptors
Within human genome, there are 48 nuclear receptors. Their gene superfamily contains numbers of ligand transcription factors for which they are categorized into two groups (i) classical endocrine hormones, and (ii) orphans. Drug induction of xenobiotics and endobiotics in liver is believed to be mediated through nuclear receptors for several factors (i.e. related to their ligand properties, binding arrangements, tissue expression) where cytochrome P450 system are incorporated. The majority of cytochrome P450 isozymes involved in drug metabolism are found in liver [7-9].
Pregnane X receptor (PXR)
One of the orphan nuclear receptors is the human pregnane X receptor (PXR). They were discovered in 1998 through similarity in structure and later on by genetic cloning in different animals. PXR are expressed in liver and gastrointestinal tract in which they regulate the transcription of genes involved in xenobiotic and endobiotic absorption, distribution, metabolism and excretion (ADME). CYP 3A4, CYP2B, CYP2C, GST, SULT, and UGT isozymes are some of the classical genes induced by PXR. Drugs that express PXR suggest their protective effect against liver toxicity (e.g. Rifampicin) [8,10,11]. In different animal and human (in vitro) models, PXR agonists suppressed liver inflammation and fibrosis. Using a rat model of ischemia reperfusion injury (IRI) has clearly showed that administration of PXR activator reduced IRI, cholestasis and fibrosis [12,13].
The objective of this study is to test the ability of each drug used in liver transplant unit to activate PXR using a luciferase reporter gene assay. Hydrocortisone and rifampicin were selected based on popularity among drugs used in liver transplant units and availability in lab.
Materials and Methods
Preparation and culture of cells
Cell line and culture media: The human liver hepatocellular carcinoma HepG2 cell line is widely used in drug liver research. HepG2 cells (obtained from Professor Matthew Wright group, Newcastle University, UK) were cultured (See culturing protocol for instructions) aseptically in Dulbecco's modified Eagle’s Medium – high glucose (DMEM) supplemented with 10% FCS (fetal-calf serum), 5% Penicillin Streptomycin and Glutamine. Cells were seeded in 6-well and 24-well plates at a capacity of 250,000 and 50,000 cell per well respectively.
Culturing protocol: To passage cells (also known as splitting), briefly rinse cell monolayer with 1X phosphate buffer saline (PBS) twice and add pre-warmed (37°C) 1% trypsin solution (5 ml).
1. Once cell layer is detached (5 to 7 min at 37°C) deactivate trypsin by adding equal volume of medium (5 ml).
2. Tap the flask once to detach cells and avoid shaking the flask while waiting.
3. Collect media/cell mixture then centrifuge at 20000 rpm for 5 minutes.
4. Remove media and leave pelleted cells.
5. Re-suspend cells in 10 ml of media and count the cells using microscope.
6. In 75 cm2 flask, transfer the cells and add the calculated volume of media.
7. Split cells at the end of the week.
8. Cultures should be incubated at 37°C in humidified atmosphere with 5% CO2.
9. Culture medium was then renewed every two to three days.
Transformation of E. coli (preparation of clone): Bacteria are used to propagate plasmids (i.e. DNA). One Shot® TOP10 chemically competent E. coli (Invitrogen, UK) used for plasmid propagation (see transformation of E. Coli, preparation of clone). These plasmids are then purified using Hispeed® Plasmid Maxi Kit ((Qiagen, UK) (For instructions check MINIPREP and MAXIPREP).
Lysogny broth (LB): Lysogny broth (LB) is used as a medium for bacterial growth. To prepare LB medium for colonization, in 500 ml bottle, 5 g tryptane, 2.5 g yeast extract, and 5 g NaCl (1.5 g/100 ml to adjust pH at 7). To prepare LB agar plates, add 7.5 g agar (1.5 g for each 100 ml). After autoclaving the mixture, let it cool then aseptically pour into round plates. Plates are now ready for colonization.
Transformation of E. coli (preparation of clone): Defrost from -80°C TOP10 competent E. coli on ice.
1. Add plasmid (0.1 μl/100 μl – 10 μl/100 μl).
2. Incubate on ice 30 min.
3. Heat-shock at 42°C for 30 seconds.
4. Place on ice for 5 min.
5. Add equal volume of lysogny broth (LB) medium (see LB), incubate at 37°C for 30 to 60 minutes with shake at 250 rpm.
6. Spread on LB agar plates (+antibiotic selection).
7. Incubate overnight at 37°C.
8. Select a single colony for miniprep and maxiprep.
Miniprep:
1. Pick a colony and inoculate 5 ml LB (+selection) and rotate overnight at 37°C.
2. Leave in fridge and use the remainder for maxiprep to isolate plasmid.
Maxiprep:
1. Pick a colony and inoculate 5 ml LB (+selection) and rotate for 8 hours at 37°C.
2. Remove 100 μl of culture (or use 100 μl from miniprep) and inoculate 100 ml LB (+selection)- incubate at 37°C with rotation.
3. Plasmid prep-use required volume of miniprep and isolate plasmid according to kit instructions.
Glycerol stocks: To prepare glycerol stock (for long term storage of plasmids), in each vial add 500 μl LB containing 30% glycerol to 500 μl of overnight culture of a single clone (autoclaved) and then keep it in -80°C freezer.
Transfection and reporter gene assay
Purified plasmids are then transfected using effectene® transfection reagent (Qiagen, UK). Transfection is made with a mixture of (ER6)3-pGL3 promoter and RLTK1 with ratio of one to ten respectively. (ER6)3-pGL3 promoter which is a construct encoding luciferase used for quantifying PXR activation [13]. While RLTK1 is a control construct encoding renilla expression not affected by anything except cell viability. Transfected cells are then treated with drug under experiment in different molar concentration. Use dimethyl sulfoxide (DSMO) as a control vehicle. For reporter gene assay follow instruction provided by Dual-Glo Luciferase system (Promega®, UK). Illuminometer or play reader devices are used to assay gene expression by quantifying the electro luminescent backlight produced. Luciferase expression for each treatment is then normalized to renilla expression and compared to those treated with DSMO.
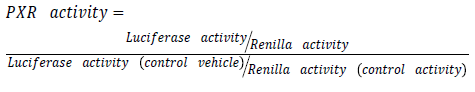
Results and Discussions
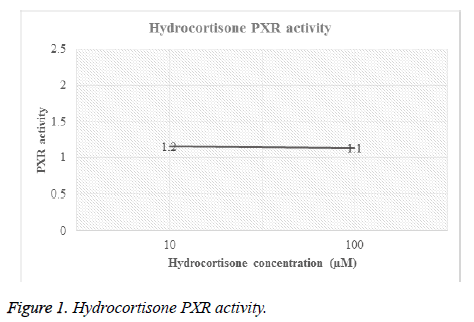
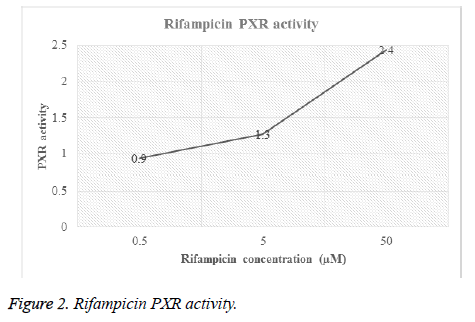
Different concentrations of hydrocortisone (HC) and rifampicin were tested for PXR activity. For each drug, transfected HEPG2 cells were treated with different molar concentrations and then luciferase activity for both firefly and renilla were recorded (Table 1, Figures 1 and 2).
| Treatment | Concentration (µM) | Firefly (F) | Illuminator reading | Mean* | PXR activity | ||||
|---|---|---|---|---|---|---|---|---|---|
| Renilla (R) | Relative light unit (RLU) | ||||||||
| Hydrocortisone | 10 | F | 2147 | 9395 | 9385 | 6440 | 9679 | 7409.2 | 1.2 |
| R | 184 | 182 | 180 | 176 | 183 | 181 | 0 | ||
| 100 | F | 2029 | 6800 | 7318 | 9146 | 11574 | 7373.4 | 1.1 | |
| R | 191 | 171 | 188 | 182 | 187 | 183.8 | 0 | ||
| Rifampicin | 0.5 | F | 4484 | 8049 | 6532 | 7017 | 4281 | 6072.6 | 0.9 |
| R | 195 | 171 | 178 | 187 | 182 | 182.6 | 0 | ||
| 5 | F | 5705 | 9506 | 8687 | 7085 | 9502 | 8097 | 1.3 | |
| R | 193 | 177 | 170 | 184 | 171 | 179 | 0 | ||
| 50 | F | 59410 | 13650 | 9382 | 11145 | 11767 | 21070.8 | 2.4 | |
| R | 471 | 190 | 182 | 184 | 189 | 243.2 | 0 | ||
| DSMO control | F | 1163 | 4834 | 6334 | 9428 | 10093 | 6370.4 | ||
| R | 187 | 170 | 171 | 184 | 186 | 179.6 | |||
Table 1. PXR activity of hydrocortisone and rifampicin.
Figures 1 and 2 showed the affinity of both hydrocortisone and rifampicin toward PXR expression. Results from rifampicin experiment support what published in the literature of their suggested protective effect against liver toxicity. Rifampicin produced a concentration-dependent increase in PXR activity. Hydrocortisone experiment needs to be tested with different concentrations to check if there is correlation between PXR expression and different doses but in general, hydrocortisone has a PXR activity.
Future Work
PXR activation in liver transplantation reduces the likelihood of graft rejection. In order to test this hypothesis research must be expanded to investigate whether patients that received PXR activating drugs were less likely to show clinical signs of graft failure than patients that received PXR non-activating drugs. “overall PXR Activation Potency” OPAP will need to be calculated for each patient, based on the drugs administered. To calculate this value, the ability of each drug used in liver transplant unit to activate PXR will be determined using a luciferase reporter gene assay. Once determined for each drug experimentally, an approximate OPAP will be calculated for each patient and compared to a variety of clinical data to see if there is correlation of OPAP with graft failure. This approach will be refined by taking into consideration the known ADME of the drugs to calculate an AUC for each drug. The OPAP will be re-calculated and compared to a variety of clinical data to see if there is a correlation of OPAP with graft failure.
Acknowledgement
The author wishes to express her sincere thanks to Professor Matthew Wright from the Institute of Cellular Medicine, Medical School, Newcastle University (Newcastle Upon Tyne, UK) for his kind help in supplying cell line and using liver research laboratory to conduct tests.
Conflict of Interest and Funding
The author declares no conflict of interest or source of funding.
References
- McCance KL, Huether SE. Pathophysiology: the biologic basis for disease in adults and children. Mosby Elsevier 2010.
- Mehrabi A. Current concepts in transplant surgery: liver transplantation today. Langenbecks Arch Surg 2008; 393: 245-260.
- World Health Organization. Transplantation. 2015. Available from: http: //www.who.int/transplantation/gkt/statistics/en/.
- Ministry of Health-Kingdom of Saudi Arabia. Statistics Book. 2013. Available from: http: //www.moh.gov.sa/en/Ministry/Statistics/book/Pages/default.aspx.
- Wood KJ, Goto R. Mechanisms of rejection: current perspectives. Transplantation 2012; 93: 1-10.
- Wiesner RH, Menon KV. Late hepatic allograft dysfunction. Liver Transpl 2001; 7: 60-73.
- Handschin C, Meyer UA. Induction of drug metabolism: the role of nuclear receptors. Pharmacol Rev 2003; 55: 649-673.
- Kliewer SA. The nuclear pregnane X receptor regulates xenobiotic detoxification. J Nutr 2003; 133: 2444-2447.
- Ogu CC, Maxa JL. Drug interactions due to cytochrome P450. Proc (Bayl Univ Med Cent) 2000; 13: 421-423.
- Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res 2002; 43: 359-364.
- Bunce CM, Campbell MJ. Nuclear receptors: current concepts and future challenges. Proteins and cell regulation. Dordrecht, Netherlands; New York: Springer 2010; 457.
- Haughton EL. Pregnane X receptor activators inhibit human hepatic stellate cell transdifferentiation in vitro. Gastroenterology 2006; 131: 194-209.
- Wallace K. The PXR is a drug target for chronic inflammatory liver disease. J Steroid Biochem Mol Biol 2010; 120: 137-148.