ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 16
The effect of XRCC3 rs861539 polymorphism on the risk of head and neck squamous cell carcinoma: a systematic review and meta-analysis
Rongxing Li, Yuqing Wen*, Zhou Lin, Yanping Zhang and Qiusheng Li
Department of Burn, Plastic Surgery, Cosmetology, Longyan First Hospital, Affiliated to Fujian Medical University, Longyan, Fujian, PR China
- *Corresponding Author:
- Yuqing Wen
Department of Burn, Plastic Surgery, Cosmetology
Longyan First Hospital
Affiliated to Fujian Medical University
Fujian, PR China
Accepted on July 06, 2017
Background: Some studies suggested that XRCC3 rs861539 polymorphism might change the risk of Head and Neck Squamous Cell Carcinoma (HNSCC). However, other studies have reported negative results. Therefore, we did this meta-analysis to investigate the role of XRCC3 rs861539 polymorphism on HNSCC risk.
Methods and Materials: Online electronic databases (PubMed, EMBASE and Wang fang database) were searched. The strength of association was assessed by calculating Odds Ratio (OR) with 95% Confidence Interval (CI).
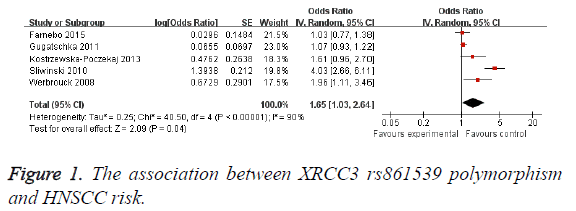
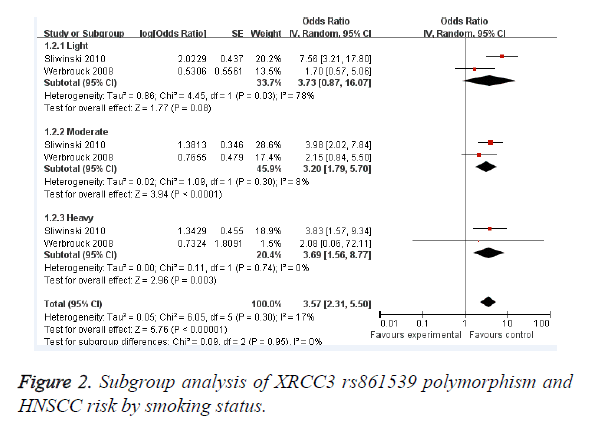
Results: A total of 5 studies with 868 cases and 1477 controls on the association between XRCC3 rs861539 polymorphism and HNSCC risk were included in this meta-analysis. Individuals with XRCC3 rs861539 polymorphism had an increased HNSCC risk (OR=1.65; 95% CI, 1.03-2.64; P=0.04). Subgroup analysis was performed according to smoking status. Light smokers with XRCC3 rs861539 polymorphism did not had an increased HNSCC risk (OR=3.73; 95% CI, 0.87-16.07; P=0.08). However, moderate smokers (OR=3.20; 95% CI, 1.79-5.70; P<0.0001) and heavy smokers (OR=3.69; 95% CI, 1.56-8.77; P=0.003) showed increased risk of HNSCC, respectively.
Conclusions: In conclusion, this study suggested that XRCC3 rs861539 polymorphism was associated with HNSCC risk.
Keywords
Head and neck squamous cell carcinoma (HNSCC), XRCC3, Meta-analysis.
Introduction
Head and Neck Squamous Cell Carcinoma (HNSCC) is one of the most common malignancies in the developing world, accounting worldwide for more than 500,000 new cases each year [1]. Surgical and chemoradiation treatments have been met with modest success. However, understanding of genetic drivers of HNSCC has been limited.
X-ray repair cross-complementing group 3 (XRCC3) belongs to the RAD51 gene family. XRCC3 involved in the Homologous Recombination Repair (HRR) of DNA double-strand break repair and cross-links [2].
Shen et al. identified a C to T substitution in exon 7 at position 18067 of XRCC3, which results in an amino acid substitution (threonine to methionine) at codon 241 [3]. Some studies suggested that XRCC3 rs861539 polymorphism might change the risk of HNSCC.
However, other studies have reported negative results [4-9]. Therefore, we did this meta-analysis to investigate the role of XRCC3 rs861539 polymorphism on HNSCC risk.
Materials and Methods
Search for publications
Online electronic databases (PubMed, EMBASE, and Wangfang database) were searched using the search terms: (XRCC3 or ‘X-ray repair cross-complementation group 3’) and (polymorphism or mutation or variation) and (head and neck squamous cell carcinoma or HNSCC). Reference lists of articles retained for review were inspected for relevant publications.
Inclusion and exclusion criteria
Two authors independently evaluated all of the studies. The study had to meet the following criteria: (1) Evaluation of XRCC3 rs861539 polymorphism on HNSCC risk; (2) Using a case-control design; (3) Genotype distributions in both cases and controls should be available for estimating an odds ratio (OR) with 95% confidence interval (CI). Studies were excluded if one of the following existed: (1) Genotype frequencies or number not offered; (2) Animal studies; (5) Editorials, reviews and abstracts. If more than one study used the same cases, the one with the most comprehensive population were included.
Data extraction and qualitative assessment
The following data was extracted: first author, year of publication, country, ethnicity, age, gender, tumor location, and the numbers of subjects. The Newcastle-Ottawa Scale (NOS) was used to evaluate the methodological quality.
Statistical analysis
Hardy-Weinberg Equilibrium (HWE) was tested using the chi-square test. A statistical test for heterogeneity was performed based on the Q statistic. The P>0.10 of the Q-test indicated a lack of heterogeneity among studies. The summary OR estimate of each study was calculated by the random-effects model. Stratified analysis was performed by smoking status. Potential publication bias was examined by funnel plot and Egger’s test. All statistical tests were performed with the software Revman version 5.1 and STATA version 11.0 (Stata Corporation, College station, TX, USA). A P value<0.05 was considered statistically significant.
Results
Characteristics of studies
A total of 5 studies with 868 cases and 1477 controls on the association between XRCC3 rs861539 polymorphism and HNSCC risk were included in this meta-analysis. All studies included Caucasian population. All the HWE results were not statistically significant. The characteristics of each study are presented in Tables 1 and 2.
| Author | Year | Country | Ethnicity | Tumor location | No. of cases | No. of controls | Age | Female (%) | HWE |
|---|---|---|---|---|---|---|---|---|---|
| Werbrouck | 2008 | Belgium | Caucasian | Mixed | 152 | 157 | 59.5 ± 10.5 | 11.5 | Yes |
| Sliwinski | 2010 | Poland | Caucasian | NA | 288 | 353 | 56 | 22 | Yes |
| Gugatschka | 2011 | Austria | Caucasian | NA | 169 | 463 | 65 ± 12 | 11.8 | Yes |
| Kostrzewska-Poczekaj | 2013 | Poland | Caucasian | Mixed | 90 | 160 | 42 ± 5.2 | 15.6 | Yes |
| Farnebo | 2015 | Sweden | Caucasian | Mixed | 169 | 344 | NA | 34 | Yes |
Table 1. Characteristics of included studies.
| Study | Selection | Comparability | Outcome | Overall quality |
|---|---|---|---|---|
| Werbrouck | 3 | 2 | 2 | 7 |
| Sliwinski | 3 | 3 | 2 | 8 |
| Gugatschka | 3 | 1 | 2 | 7 |
| Kostrzewska-Poczekaj | 3 | 3 | 2 | 8 |
| Farnebo | 3 | 2 | 3 | 8 |
Table 2. Quality scores of studies using Newcastle-Ottawa Scale.
Meta-analysis results
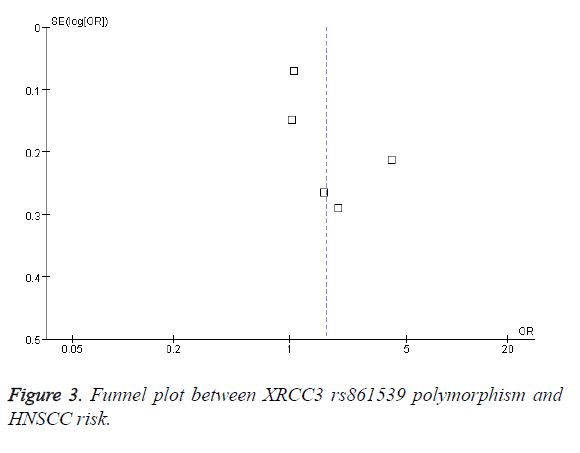
As shown in Figure 1, individuals with XRCC3 rs861539 polymorphism had an increased HNSCC risk (OR=1.65; 95% CI, 1.03-2.64; P=0.04). Subgroup analysis was performed according to smoking status. Light smokers with XRCC3 rs861539 polymorphism did not had an increased HNSCC risk (OR=3.73; 95% CI, 0.87-16.07; P=0.08) (Figure 2). However, moderate smokers (OR=3.20; 95% CI, 1.79-5.70; P<0.0001) and heavy smokers (OR=3.69; 95% CI, 1.56-8.77; P=0.003) showed increased risk of HNSCC, respectively. Funnel plot was performed to assess the publication bias of literatures. The shape of the funnel plot showed symmetry (Figure 3). Egger’s test found no evidence of publication bias (P=0.8) (Table 3).
| OR (95% CI) | P | I2 | |
|---|---|---|---|
| Overall | 1.65 (1.03-2.64) | 0.04 | 90 |
| Smoking status | |||
| Light smokers | 3.73 (0.87-16.07) | 0.08 | 78 |
| Moderate smokers | 3.20 (1.79-5.70) | <0.0001 | 8 |
| Heavy smokers | 3.69 (1.56-8.77) | 0.003 | 0 |
Table 3. Results of the meta-analysis.
Discussion
To our knowledge, this was the first meta-analysis to investigate the role of XRCC3 rs861539 polymorphism on HNSCC risk. This meta-analysis included 5 studies with 868 cases and 1477 controls. We found that individuals with XRCC3 rs861539 polymorphism had an increased HNSCC risk. Subgroup analysis was performed according to smoking status. Light smokers with XRCC3 rs861539 polymorphism did not have an increased HNSCC risk. However, moderate smokers and heavy smokers showed increased risk of HNSCC, respectively.
Carriers of the variant allele of XRCC3 rs861539 polymorphism had different DNA adduct levels in lymphocyte DNA, and the Met variant was significantly associated with higher DNA adduct levels, indicating that this polymorphism was associated with the DNA repair capacity [9]. This polymorphism has been studied in many other diseases. Lu et al. concluded that the XRCC3 Thr241Met polymorphism is associated with an increased risk of thyroid cancer in the overall population [10]. Chai et al. suggested that XRCC3 Thr241Met polymorphism might be associated with breast cancer risk, especially in Asian populations [11]. Cheng et al. suggested that XRCC3 gene rs861539 polymorphism was associated with the risk for gastric cancer in Asian populations [12]. Bei et al. did not find a significant correlation between XRCC3 Thr241Met polymorphism and lung cancer risk [13]. Feng et al. also suggested that XRCC3 T241M polymorphism did not confer glioma risk [14].
Some limitations of this meta-analysis should be acknowledged. First, the number of published studies was not sufficiently large for a comprehensive analysis. Second, the sample size was relatively small. Third, there was clinical and statistical heterogeneity among included studies
In conclusion, this meta-analysis suggested that XRCC3 rs861539 polymorphism was significantly associated with HNSCC risk.
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer 2001; 94: 153-156.
- Brenneman MA, Weiss AE, Nickoloff JA, Chen DJ. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat Res 2000; 459: 89-97.
- Shen MR, Jones IM, Mohrenweiser H. Non conservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res 1998; 58: 604-608.
- Werbrouck J, De Ruyck K, Duprez F, Van Eijkeren M, Rietzschel E, Bekaert S, Vral A, De Neve W, Thierens H. Single-nucleotide polymorphisms in DNA double-strand break repair genes: association with head and neck cancer and interaction with tobacco use and alcohol consumption. Mutat Res 2008; 656: 74-81.
- Sliwinski T, Walczak A, Przybylowska K, Rusin P, Pietruszewska W, Zielinska-Blizniewska H, Olszewski J, Morawiec-Sztandera A, Jendrzejczyk S, Mlynarski W, Majsterek I. Polymorphisms of the XRCC3 C722T and the RAD51 G135C genes and the risk of head and neck cancer in a Polish population. Exp Mol Pathol 2010; 89: 358-366.
- Gugatschka M, Dehchamani D, Wascher TC, Friedrich G, Renner W. DNA repair gene ERCC2 polymorphisms and risk of squamous cell carcinoma of the head and neck. Exp Mol Pathol 2011; 91: 331-334.
- Kostrzewska-Poczekaj M, Gawęcki W, Illmer J, Rydzanicz M, Gajecka M, Szyfter W, Szyfter K. Polymorphisms of DNA repair genes and risk of squamous cell carcinoma of the head and neck in young adults. Eur Arch Otorhinolaryngol 2013; 270: 271-276.
- Farnebo L, Stjernström A, Fredrikson M, Ansell A, Garvin S, Thunell LK. DNA repair genes XPC, XPD, XRCC1, and XRCC3 are associated with risk and survival of squamous cell carcinoma of the head and neck. DNA Repair 2015; 31: 64-72.
- Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S, Piazza A, Vineis P. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis 2001; 22: 1437-1445.
- Lu W, Wu G, Zhang B. Association between X-Ray cross-complementing group 3 (XRCC3) Thr241Met polymorphism and risk of thyroid cancer: A meta-analysis. Med Sci Monit 2015; 21: 3978-3985.
- Chai F, Liang Y, Chen L, Zhang F, Jiang J. Association between XRCC3 Thr241Met polymorphism and risk of breast cancer: meta-analysis of 23 case-control studies. Med Sci Monit 2015; 21: 3231-3240.
- Cheng S, Wang L, Wang L, Wang Z. Association of XRCC3 gene rs861539 polymorphism with gastric cancer risk: evidence from a case-control study and a meta-analysis. Int J Clin Exp Pathol 2015; 8: 1911-1919.
- Bei L, Xiao-Dong T, Yu-Fang G, Jian-Ping S, Zhao-Yu Y. DNA repair gene XRCC3 Thr241Met polymorphisms and lung cancer risk: a meta-analysis. Bull Cancer 2015; 102: 332-339.
- Feng Y, Zeng M, Xu Q. Association between XRCC3 T241M polymorphism and glioma risk: a meta-analysis. Tumour Biol 2014; 35: 5589-5592.