ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2016) Volume 27, Issue 4
The research of the relationship between new single nucleotide polymorphism G5508A in the SEPT12 gene and azoospermia
Changqing An1#, Lijuan Han2#, Guiying Yu3#, Pan Li4# and Yanzhi Xia4*
1Department of Paediatrics, Hubei Maternal and Child Health Hospital, Wuhan, Hubei, China
2Department of Dermatology, the First Hospital of Wuhan, Wuhan, Hubei, China
3Hand and Foot Microsurgery, the Second Hospital of Jingzhou, Jingzhou, Hubei, China
4College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, Hubei, China
#These authors contributed equally to this work
- *Corresponding Author:
- Yanzhi Xia
College of Life Science and Technology
Huazhong University of Science and Technology
China
Accepted date: April 06, 2016
The objective of the present study is to investigate the association between SEPT12 gene variation and the risk of azoospermia. In the current study, 100 infertile patients with azoospermia and 100 normal controls were analyzed for genetic alterations in the active site coding region of SEPT12, using PCRSequencing technique and mutational analysis. Genotype analysis indicated that G5508A polymorphic SEPT12 alleles were distributed in three peaks of frequency in diseases group (GG, GA and AA) and two peaks in control group (GG and GA). The frequencies of the G5508A allele and the genotype were significantly difference between the azoospermic patients and controls (p ≤ 0.05 by chi-square test). This finding suggests that G5508A variant in the SEPT12 gene may associate with the increased susceptibility to azoospermia.
Keywords
Male infertility, Azoospermia, Polymorphism, SEPT12.
Introduction
Subfertility is of major clinical socioeconomic concern. Around 10% to 15% of the couples worldwide are affected by reduced fertility, and the defects can be traced to the men in roughly half of the cases [1]. The pathology of male infertility includes anatomic defects, gametogenesis dysfunction, endocrinopathies, immunologic problems, ejaculatory failure, environmental exposures, and gene mutations [2,3]. In about 35% of the couples seeking medical attention, the male partners are diagnosed with spermatogenic failure, defined as one or more semen parameters falling below the WHO cut-off for normozoospermia [4]. Of the most severe forms of male infertility are azoospermia which is defined as complete absence of sperm from ejaculate, and asthenospermia, which means having defects in sperm motility [1].
Spermatogenesis is governed by the parallel and serial actions of thousands of genes. Alterations in any of them or their expression may cause male infertility [5]. In fact, only a handful of genetic alterations have clearly been shown to cause spermatogenic failure [6].
Septins are a highly conserved polymerizing GTP-binding protein family. The functions of SEPTs are diverse, and include establishing cell polarity, cytoskeletal remodeling, membrane compartmentalization, cell cycle progression, and vesicle trafficking. Among the 14 members of the septin gene family of mammalian species, several members are expressed ubiquitously, whereas others are expressed only in welldifferentiated cells (e.g., neurons or male germ cells) [7]. Since 1997, 14 members of septin proteins have been characterized for humans (SEPT1-SEPT14), some of which are tissuespecific. All of the 14 genome-mapped human septin genes encode a highly conserved central GTP binding/hydrolysis domain, which is very critical in GTPase signaling properties, as well as oligomerization between septins and other filamentous proteins. This functional domain consists of three distinct amino acid sequence elements, G1, G3 and G4, which share sequence identity to the well-characterized Ras GTPase family [8].
Among all septins, SEPT12 is dominantly expressed in testis tissue of adults, with an established essential role in annulus structure of mature sperm [9]. Recently, it has been shown by investigators that high levels of SEPT12 mRNA is observable exclusively in the testis of sexually mature males (human and mouse), while this mRNA was not detectable in men with sterility resulting from inability to produce mature spermatozoa. So, it has been considered that SEPT12 is crucial for the process of spermatogenesis in mammals [10].
In this study, we tried to monitor genetic variations of SEPT12 gene in a population of male infertile Chinese with nonobstructive azoospermia, in order to investigate the association between SEPT12 gene variation and azoospermia.
Materials and Methods
Patients
The study population consisted of 100 azoospermic male (age: 35.7 ± 7.8) who were referred to Hubei Maternal and Child Health Hospital from 1/1, 2014 to 31/12, 2014. In addition, 100 male (age 31.4 ± 4.2) with normal spermograms were analyzed as control (Table 1). Patients with hypogonadotropic hypogonadism, cryptorchidism, orchitis, ejaculatory duct obstruction, and men with microdeletions of the long arm of Ychromosome or karyotype abnormalities were excluded from the study. The classification of men into the normozoospermic and azoospermic groups was according to World Health Organization criteria [1]. Informed consent was granted from all individuals in the study.
Genomic DNA was extracted from peripheral blood specimens by a QIAamp DNA minikit (Takara Japan), according to the instructions provided by the manufacturer.
NCBI SNP and 1000Genome phase I annotation of SNP site
The total depth of mapped sequence reads, the average mapping quality score and the fraction of reads with mapping quality zero (meaning that this read maps equally well to more than one location in the genome) were tabulated from 1,103 bam files in the 1000 Genomes Phase 1 low coverage data release. This combines low coverage of the whole genome sequence information from 1,092 individuals, giving a genome wide average total depth of coverage of 5,132 reads. Both "pilot" and "strict" tracks are .bed file conversions of the "pass" regions from FASTA mask files.
The "pilot" criteria require a depth of coverage between 2,566 and 10,264 inclusive (between one-half and twice of the average depth), and that no more than 20% of covering reads have mapping quality zero. These are equivalent to the criteria used for the analyses in the 1000 Genomes Pilot paper (2010). The "strict" criteria require a depth of coverage between 2,566 and 7,698 inclusive, no more than 0.1% of reads with mapping quality zero, and an average mapping quality of 56 or greater. This definition is quite strict and focuses on the most unique regions of the genome. In the regions which passed the strict criteria, only 2% of sites called in an initial analysis were rejected as likely false positives by VQSR. Since approximately one-half of the 1000 Genomes Project individuals are males, the depth of coverage is generally lower on X chromosome than Y chromosome. The coverage thresholds on X chromosome were adjusted by a factor of 3/4, and on Y chromosome by a factor of 1/2. The "pilot" criteria were not evaluated for Y chromosome.
Next generation sequencing ampliseq analysis
The Ion Torrent Library was prepared via an Ion AmpliSeq Kit and an Ion AmpliSeq Cancer Panel v2 (Life Technologies) for the remaining samples. The latter allows for simultaneous amplification of 207 amplicons in hotspot areas of 50 genes (SEPT12 is included with the same PCR amplification primer set same as follows the forward primer is 5’- AAGCTGCAAGGTAGCAACAGC-3’, the reverse primer is 5’-CTGCAGCTTTTTCTCTAGGG-3’) [3,5]. PCR was performed in 17 cycles for FF samples and in 20 cycles for FFPE samples. Samples were barcoded by anIonXpress Barcode Adapters (Life Technologies) to allow for the discrimination among the samples within a NGS run. The DNA concentration of the samples within one sequencing run was normalized by a Qubit 2.0 fluorometer (ThermoFisher Scientific) or an Ion Library Equalizer kit. The Ion AmpliSeq Library Kit 2.0 (Life Technologies) was used for library preparation. The library was mixed with Ion Sphere Particles (ISPs). The subsequent emulsion PCR and enrichment were performed by an Ion PGMTM Template OT2 200 Template Kit and an Ion One Touch 2 instrument (Life Technologies). Sequencing was performed by an Ion PGMTM Sequencing 200 kit v2 with an Ion 316TM or 318TM chip (Life Technologies) (the maximum number of samples on 316 and 318 chip were 6 and 12 respectively). After sequencing, the raw fastq famat was genetated and mapped to the human genome (hg19). The SEPT12 regions were selected. And IGV was used to visualize SNP condition.
PCR-Sequencing analysis
For genetic analysis, the coding region of G1, G3 and G4 functional domains of SEPT12 were screened by Polymerase Chain Reaction (PCR)-directed sequencing, the forward primer is 5’-AAGCTGCAAGGTAGCAACAGC-3’, the reverse primer is 5’-CTGCAGCTTTTTCTCTAGGG-3’ [3,5].
To analyze the aforementioned DNA sequences, PCR was performed in a final volume of 25 μl, consisting of genomic DNA (10 ng), dNTP (0.32 mM each), each primer (0.2 μM), 0.2 μM Taq polymerase (0.625 IU) and a reaction buffer containing MgCl2. PCR was performed as follows initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 45 sec, annealing at 60°C for 45 sec and extension at 72°C for 45 sec. The PCR products were confirmed by running on 1.5% agarose gel (Figure 1), and were applied for sequencing by an ABI 3500Dx automated DNA sequencer.
qRT-PCR assay of SEPT12 expression level
Six patients and six normal samples were selected. All the blood samples were 200 μl with 200 μl Trizol (Invitrogen). After lysis, the chromfol and phenole were used for total RNA extracted. A Fermentas K1622 (ThermoFisher) kit was applied for the reverse transcription of total RNA to cDNA. After cDNA was acquired and quatitated, qPCR methold was used to detecte the expression level of SEPT12 with primer set.
Statistical analysis and odds ratio of SNP
Chi-squared test was used to compare the frequencies of allele frequency. The odds ratio (a probability value) of less than 0.05 was considered to be statistically significant. The OR within one group was compared to the OR within another group, in order to associate an allele or a genotype to a phenotype (usually a trait or a disease).
Typically, the carriers of a less common allele or genotype are compared to people with two copies of the most common allele. An OR of 1.0 means that the DNA variant has no effect on the odds of having the disease. An OR above 1.0 indicates a statistical association between the variant and the disease, while below 1 indicates a weak association (risk).
The variants with a modest Mendelian effect have an OR of 3 or greater, where the highly penetrant variants can have even higher ORs. Although commonly reported for SNP associations with diseases, be wary of the tendency of the OR to overemphasize the effect of a SNP. This is particularly true in cases where the SNP in question is quite rare in both groups, and/or the group sizes themselves are rather small.
Results
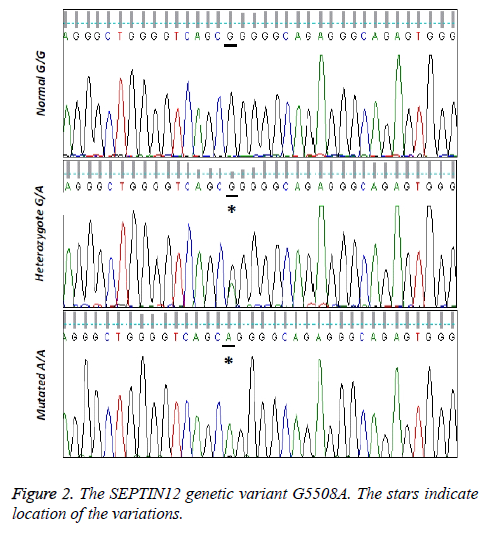
The sequence analysis data reveals the nucleotide transition G5508A in SEPT12 gene of the patients in comparison with normal controls (Figure 2). As indicated in Table 1, 64 samples among the 100 azoospermic patients are normal homozygous (GG). 28 of them have the heterozygous (GA) mutation and 8 individuals are observed with both alleles mutated (AA). Statistical analysis of the data shows that the G5508A variation of SEPT12 gene reveals significantly different genotype distributions between the normal control group and the patients groups (p<0.05). In addition, the frequency of the normal G allele is significantly higher in the control group than in the patients groups (p<0.05).
| Normal | Azoospermia | |
|---|---|---|
| Genotype-wise comparison | ||
| GG | 91 | 64 |
| GA | 9 | 28 |
| AA | 0 | 8 |
| Allele-wise comparison | ||
| G | 191 | 156 |
| A | 9 | 44 |
Table 1. Distribution of the allelic alteration of G5508A between azoospermic patients and normal controls.
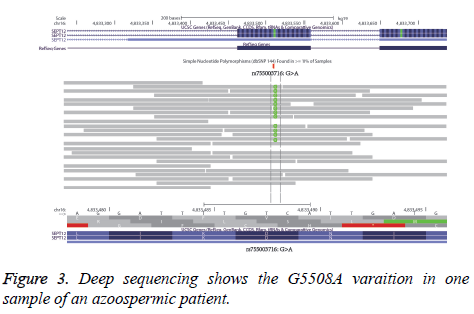
The NGS deeply sequences the three samples of the patients, and checks the G5508A variation of the SEPT12 condition. The IGV program shows the number reads of G and the number of A in Figure 3. According to the data of one patient, 70 reads shows GG variation and 34 reads shows GA variation. This ratio is close to 64/28, the results of the sequence analysis mentioned above.
Six samples of patients’ and normal people’s blood were taken for gene expression validation. After qRT-PCR assay, we found the expression of gene SEPT12 did not significantly change. It shows that the SNP change is not able to affect the gene expression.
In the NCBI SNP database, the 1000 genome project and phase I and phase II data have been released since 2010s. We annotate the SETIN12 genetic variant G5508A to find the relevant serial number and annotation information. It was found out that rs755003716 is this SNP serial number and conducts D [Asp] → G [Gly] of NP_653206.2 (SETIN12 protein). It strongly suggests that G5508A changes the protein amino acid and causes the disease.
Discussion
There are several reports on novel genes involved in spermatogenesis, in the way that their down-regulation in the testes tissues of infertile men has been identified by high throughput expression analyses [11,12]. SEPT12 is one of these genes whose lower expression in the testicular biopsies of infertile men is strongly related with azoospermia.
In this study, a SEPTIN12 genetic variant G5508A was found to be significantly associated to male infertility with distinctive sperm pathology. The allele frequency of G5508A in SEPT12 gene is significantly different between Chinese patients with azoospermia and the controls with normal spermograms (P<0.05). There was a difference in age between the patient group (35.7 +/- 7.8) and the control group (31.4 +/- 4.2) (P<0.01). But the age can not alter the basic group of SEPT12 gene.
These results are not yet conclusive that SEPT12 gene mutations are the direct reason for azoospermia, as the sample size of the patients in this analysis is not big enough [13]. However, our preliminary observation among these Chinese patients suggests that this G>A alteration may be one of the important factors to disrupt spermatogenesis.
Six samples of patients’ and normal peoples’ blood were taken for gene expression validation. The qRT-PCR assay did not reveal any significant change on the expression of gene SEPT12. This demonstrates that the SNP change does not affect the gene expression.
In the NCBI SNP database, we annotate the SETIN12 genetic variant G5508A to find the relative serial number and annotation information. We found that rs755003716 is this SNP serial number and conduct D [Asp] → G [Gly] of NP_653206.2 (SETIN12 protein). It is a strong indication that G5508A may change the protein amino acid and cause the disease.
According to the recent reports that some SEPT12 SNPs may predispose men to spermatogemic failure [4,14], the current data propose that the novel G5508A polymorphism can be considered as a biomarker for idiopathic male infertility. Our results may provide a new insight at molecular level for nonobstructive azoospermia. It is still to be confirmed if the link between this variant and azoospermia holds true for patients among other ethnic groups.
In conclusion, we suggest that the novel G5508A polymorphism in SEPT12 gene may predispose Chinese men to a defect in spermatogenesis.
References
- WHO (1999) WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction.
- Sadeghi-Nejad H, Farrokhi F. Genetics of azoospermia: current knowledge, clinical implications, and future directions. Part I. Urol J 2006; 3: 193-203.
- Aryanpur M, Tarahomi M, Sharifi H, Heydari G, Hessami Z, Akhoundi M. Comparison of spermatozoa quality in male smokers and nonsmokers of Iranian infertile couples. Int J Fertil Steril 2011; 5: 152-157.
- Toshinobu Miyamoto, Akira Tsujimura, Yasushi Miyagawa. Single nucleotide polymorphisms in the SEPTIN12 gene maybe associated with azoospermia by meiotic arrest in Japanese men. J Assist Reprod Genet 2012; 29: 47-51.
- Gava MM, Chagas Ede O, Bianco B, Christofolini DM, Pompeo AC, Glina S. Methylenetetrahydrofolate reductase polymorphisms are related to male infertility in Brazilian men. Genet Test Mol Biomarkers 2011; 15: 153-157.
- Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online 2008; 16: 504-513.
- Hall PA, Jung K, Hillan KJ. Russell SE. Expression profiling the human septin gene family. J Pathol 2005; 206: 269-278.
- Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol 2008; 9: 478-489.
- Steels JD, Estey MP, Froese CD, Reynaud D, Pace-Asciak C, Trimble WS. Sept12 is a component of the mammalian sperm tail annulus. Cell Motil Cytoskeleton 2007; 64: 794-807.
- Lin YH, Lin YM, Wang YY, Yu IS, Lin YW, Wang YH. The expression level of septin12 is critical for spermiogenesis. Am J Pathol 2009; 174: 1857-1868.
- Yung-Che Kuo, Ying-Hung Lin, Hau-Inh Chen. SEPT12 mutations cause male infertility with defective sperm annulus. Human Mutation 2012; 33: 710-719.
- Maryam S, Mahnaz A, Marjan S. New single nucleotide polymorphism G5508A in the SEPT12 gene may be associated with idiopathic male infertility in Iranian men. Iran J Reprod Med 2015; 13: 503-506.
- Burton PR, Clayton DG, Cardon LR. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661-678.
- Miyakawa H, Miyamoto T, Koh E. Single-nucleotide polymorphisms in the SEPTIN12 gene may be a genetic risk factor for Japanese patients with Sertoli cellonly syndrome. J Androl 2012; 33: 483-487.