ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2016) Volume 27, Issue 3
Visualization and quantification of SPIO nanoparticles in intracellular spaces of macrophages for nanomedicine applications.
Department of Biomedical Technology, College of Applied Medical Sciences, King Saud University, Riyadh 11433, Saudi Arabia
- *Corresponding Author:
- Ali Saad
Department of Biomedical Technology
King Saud University, Saudi Arabia
Accepted Date: February 24, 2016
This work focuses on the visualization and quantification of Super-paramagnetic iron oxide nanoparticles (SPIO-NPs) inside macrophages using transmission electron microscope (TEM) images. Macrophages have been extensively used as probes for cell and subcellular structure labeling, as well as for drug and gene delivery. In order to track the macrophages movement inside the body, SPIO-NPs are introduced to help in the visualization of macrophages by MRI imaging and then to control process of the drug and gene delivery. The mechanism of interaction between macrophages and SPIO-NPs is still not fully understood by scientists. Estimating the quantity of NPs inside microphages will certainly improve the understanding of this mechanism. In this work a 3D reconstruction and visualization, using transmission electron microscopy (TEM), of macrophages filled with SPIO-NPs is being developed. An estimation of the quantities of the NPs inside macrophages has been achieved. The visual interaction between macrophages and SPIO-NPs had been discovered.
Keywords
Nanoparticles, Macrophages, Quantification, Visualization, TEM, 3D Reconstruction
Introduction
The rapid advances in nanotechnology and the ever-decreasing size of features in the biological sciences result in the need for advanced characterization with high spatial resolution in two and three dimensions. Electron microscopy, using tomography to reconstruct a three-dimensional nature of an object from a tilt series of two-dimensional projections (images), allows more pertinent quantitative analysis. Electron tomography offers an intermediate resolution (of about 1 nm) with a field of view of hundreds of nanometer, making it ideal for the characterization of many nano-scale objects. It is the most widely applicable method for obtaining three-dimensional information by electron microscopy.
Once administered in the body, nanoparticles will be essentially taken up by the reticulo-endothelial system (RES) consisting mainly of macrophages [1], which are involved in the coordination of immune responses, the elimination of pathogens, and the control of tissue homeostasis.
Certain studies focus on the understanding of the undesirable, unintended or toxic effects of NPs once administrated to the human body. In order to understand side effects that may occur in vivo during transport of NPs to the area of interest, a recent study [2] tried to understand and exploit NPs interaction with the blood vessel and blood. Another study [3] investigated the physico-chemical properties of NPs in order to understand the toxic effect of nanomaterials on the cell biology. Also, the phenomena of agglomeration of NPs in cells may affect the potential of drug delivery, diagnostic imaging or increase the toxic potential. A rapid screening method to evaluate agglomeration of NPs in the blood is proposed in [4]. Epithelial cell sheet showed slower migration after exposure to TiO2 and SiQ2 NPs and significantly impaired wound healing capability [5].
With their fascinating size-dependent magnetic properties and their relative biocompatibility, magnetic Superparamagnetic iron oxide (SPIO) nanoparticles have been extensively used as probes for cell and subcellular structure labeling, as well as for drug and gene delivery. They were commonly applied for different biomedical applications, such as magnetic resonance imaging (MRI) and drug delivery to living cells [6,7].
Non-invasive imaging of macrophage activity using SPIO nanoparticles as contrast agent has raised increasing interest in the diagnosis of diseases involving inflammation, infections and tissue degeneration, making them attractive vehicles to deliver contrast agents for diagnostic or drugs for therapeutic purposes [8]. Inside the body, different environmental conditions will orient macrophages to have different actions or polarization states [9,10]. Classically activated or M1 macrophages have a pro-inflammatory action. In contrast, alternatively activated or M2 macrophages, have an immunomodulating activity and promote wound healing and angiogenesis [11].
Therefore, a better understanding of the mechanisms of interaction between nanoparticles and macrophages would significantly advance our control of dynamic processes in nanomedicine.
In this paper, a 3D reconstruction and visualization of macrophages with SPIO-NPs at a sub-cellular scale is seen. Moreover, a method of quantification of NPs uptake by macrophages is proposed. This will help investigators to better estimate the amount of NPs absorbed by macrophages. This visualization and quantification serves the purpose of screening the agglomeration of NPs, the control of drug and gene delivery process as well as improve the diagnosis and treatment of different diseases by MRI imaging [12,13].
Materials and Methods
Macrophages polarization and labeling
Bone marrow (BM) derived M1 and M2 macrophages (BMDM) were obtained as previously reported [14].
Dextran coated iron oxide nanoparticles functionalized by the addition of Polyethylene Glycol (PEG) were preferred for efficient macrophages labeling [15,16]. It was previously reported that these nanoparticles showed an enhanced labeling efficiency of macrophages with a better biocompatibility [17]. These nanoparticles have a dextran coating of 40,000 g/mol and a PEG length of 300 g/mol for a total diameter of 100 nm with an iron oxide crystallite diameter of 10-13 nm according to the supplier specifications.
M1- and M2-polarized macrophages were incubated with either carboxyl or amine modified PEGylated SPIO nanoparticles (SPIO-PEG-COOH or SPIO-PEG-NH2 respectively) in serum-free RPMI medium to avoid proteins interactions with the uptake mechanism [18] as the colloidal stability of particles in protein-rich media can be influenced by interface characteristics [19]. The incubation step was followed by 4-5 hours in SPIO-free culture medium to allow sufficient time for iron oxide internalization before fixation for TEM tomographic analysis.
Sample preparation and TEM tomography imaging
M1 and M2 Macrophages were cultured and labeled in Permanox dishes to allow direct processing of cells without the need for scraping. They were fixed with 2.5% glutaraldehyde in 0.05 M sodium cacodylate buffer (pH 7.4), post-fixed with 2% osmium tetroxide, subsequently dehydrated in ascending grades of ethanol and finally embedded in Epon resin. Ultrathin sections (70 nm) were imaged with tilt series (-60° with 2° increment step size) at resolution of 9.7Å/pixel using a Hitachi H7500 transmission electron microscope operating at 80 kV (National Center for Macromolecular Imaging, Baylor College of Medicine, TX, USA) with a time of exposure of 1s and a defocus set to 0 μm. Golden particles of size 15 nm were used as fiducial seeds for alignment purpose.
3D reconstruction and visualization
The 3D reconstruction uses TomoJ software [20] for transmission electron tomography. TomoJ combines multiple tomographic volumes and includes the most recent algorithms for volume reconstructions used in three-dimensional electron microscopy. It also includes the algebraic reconstruction technique and simultaneous iterative reconstruction technique, as well as the commonly used approach of weighted backprojection. The workflow in [21,22] was used for microphages 3D reconstructions. 3D Slicer software was used for visualization. 3D Slicer is a free open source software platform for medical image processing and 3D visualization of image data.
Results and Discussion
Figure 1 show three samples of tomographic tilt-series of the macrophage with SPIO1-m0.
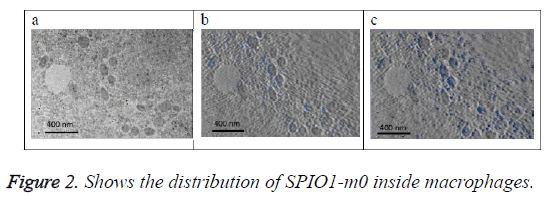
Figure 2a shows the original image (projection). Figure 2b shows the SPIO in blue color that exists on this section only determined from 3D model of the SPIO in macrophages. Figure 2c shows a projection of 3D reconstructed model of the SPIOs in macrophages at a direction (90°) perpendicular to the slice plan. SPIOs nanoparticles are shown in blue color in the images at different locations.
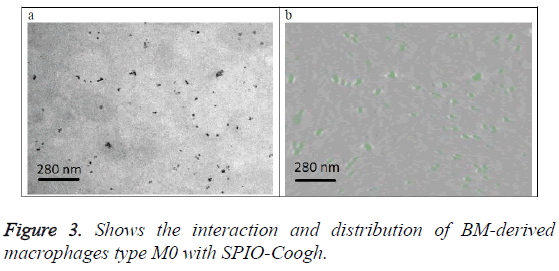
Figure 3a shows the original image (projection). Figure 3b shows a slice from the 3D volume of the SPIO-Coogh macrophages, SPIOs are in green color. The slice is at the same location as the section of figure 3a.
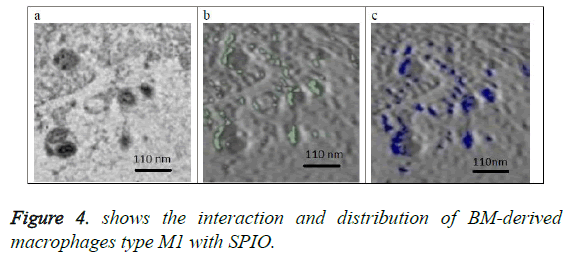
Figure 4a shows the original image (projection) located at the center of the 3D volume of the SPIO in the macrophage. Figure 4b shows in green the SPIOs in this section only. Figure 4c shows a slice from the volume of macrophages with same location as the section in figure 4b. SPIOs are shown in blue color on the slice of figure 4c.
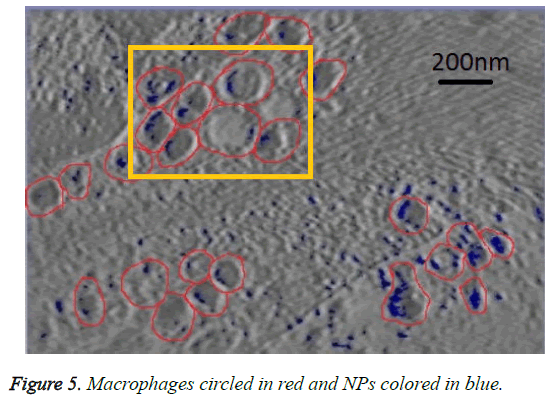
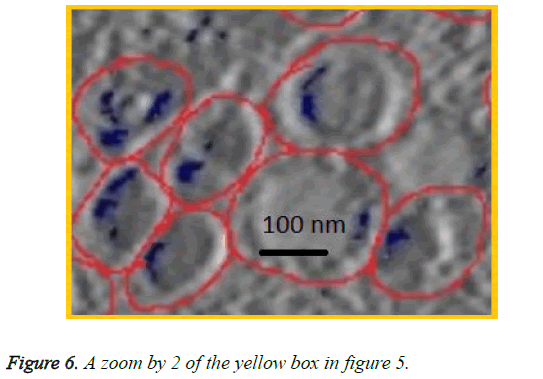
We can see in figures 5 and 6 that macrophages inside the red circles are in a different dimension. The blue dots in figure 6 represent the SPIO NPs .The figure also shows different amounts of NPs in blue color inside each macrophage as well as certain macrophages have no NPs. This view shows that the SPIO are inside the macrophages. From this result it is easy to assess the quantity of SPIOs engulfed by the macrophages by counting the blue dots inside the macrophage, and then making an overall estimation of the number of NPs inside macrophages.
The resolution of the TEM images is about 10 angstrom per pixel (1 nm) and the average diameter of NPs used is about 11 nm. Counting the blue pixels in the encircled macrophage in the slice provides an estimation about the surface covered by NPs inside this macrophage. If the blue surface is divided by the average surface of one NP, this provides an estimation of the total number of NPs inside the encircled macrophage. The surface of one SPIO-NP is estimated by πd2/4, with average diameter of 11 nm.
Assuming that the number of NPs outside macrophages is negligible, simply counting the total number of NPs inside the slice and dividing it by the total number of macrophages in the same slice provides an estimated average number of NPs in each macrophage.
Visualization of the 3D distribution of NPs will help to locate the macrophages, measure their dimensions and counts them. From the whole set of blue dots we can estimate the average number of NPs absorbed by each macrophage. The NPs helps in seeing the macrophages locations using MRI images. Macrophages are used as vehicle for drug delivery and NPs can helps to detect the macrophage numbers and locations inside the target organ using MRI imaging. This visualization and quantification improves the accuracy of drug delivery by estimating the amount of drug received by the site of interest (inflammation or cancer). It can also help in the diagnosis and therapy procedures of the region of interest in nanomedicine.
Seeing the NPs inside macrophages in 3D and knowing their approximate amount will certainly help in reduction of the undesirable effect of NPs as well as in the control of drug delivery and imaging processes for nanomedicine applications.
Conclusion
This work has shown that macrophage SPIO interaction can be visualized in 3D. It also shows that most of the SPIOs are incorporated into the macrophages. Hence, this study could help in assessing the quantity of SPIOs inside the macrophage in 3D space. This provides researchers a more accurate framework for drug delivery, diagnosis imaging or other nanomedicine applications.
Acknowledgements
The author extends his appreciation to the College of Applied Medical Sciences Research Centre and the Deanship of Scientific Research at King Saud University for funding this research. I also thank Prof. W. Chiu, Dr. Q. Zhang, and Dr. A. Al-Faraj for sample preparation and TEM imaging.
References
- Lartigue L, Wilhelm C, Servais J, Factor C, Dencausse A, Bacri JC, Luciani N, and Gazeau F. Nanomagnetic sensing of blood plasma protein interactions with iron oxide nanoparticles: impact on macrophage uptake. ACS Nano 2012;6: 2665-2678.
- Magdiel S, Chor YT, Dominic D, Roland HS, David TL. Understanding and Exploiting Nanoparticles' Intimacy with the Blood Vessel and Blood. Chemical Society Reviews 2015; 44: 8174-8199.
- Tay CY, Magdiel IS, Jianping X, Wolfgang JP, David TL. Back to basics: Exploiting the innate physico-chemical characteristics of nanomaterials for biomedical applications. Advanced Functional Materials 2014; 24: 5936-5955
- Samir VJ, Haiou Q, Thilak M, Taylor MI, Rongrong W, Feng W, Paul CH, Jingyi C, Yongbin Z. Rapid determination of plasmonic nanoparticle agglomeration status in blood. Biomaterials 2015; 51: 226-237.
- Chor YT, Pingqiang C, Magdiel IS, Wanru F, Lay PT, Catherine H, Xiaodong C, David TL. Nanoparticles strengthen intracellular tension and retard cellular migration. Nano Letters 2014; 14: 83-88.
- Xie J, Huang J, Li X, Sun S, Chen X. Iron oxide nanoparticle platform for biomedical applications. Curr Med Chem 2009; 16: 1278-1294.
- Issa B, Obaidat IM, Albiss BA, Haik Y. Magnetic nanoparticles: surface effects and properties related to biomedicine applications. Int J MolSci 2013; 14: 21266-21305.
- Beduneau A, Ma Z, Grotepas CB, Kabanov A, Rabinow BE, Gong N, Mosley RL, Dou H, Boska MD, Gendelman HE. Facilitated monocyte-macrophage uptake and tissue distribution of superparmagnetic iron-oxide nanoparticles. PLoS One 2009; 4: e4343.
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8: 958-969.
- Van Ginderachter JA, Movahedi K, HassanzadehGhassabeh G, Meerschaut S, Beschin A, Raes G, De Baetselier P. Classical and alternative activation of mononuclear phagocytes: picking the best of both worlds for tumor promotion. Immunobiology 2006; 211: 487-501.
- Ho VW, Sly LM. Derivation and characterization of murine alternatively activated (M2) macrophages. Methods MolBiol 2009; 531: 173-185.
- Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater 2009; 8: 543-557.
- Hillaireau H, Couvreur P. Nanocarriers? entry into the cell: relevance to drug delivery. Cellular and Molecular Life Sciences 2009; 66: 2873-2896.
- Al Faraj A, Luciani N, Kolosnjaj-Tabi J, Mattar E, Clement O, Wilhelm C, Gazeau F. Real-time high-resolution magnetic resonance tracking of macrophage subpopulations in a murine inflammation model: a pilot study with a commercially available cryogenic probe. Contrast Media Mol Imaging 2013; 8: 193-203.
- Tang KS, Shapiro EM. Enhanced magnetic cell labeling efficiency using -NH2 coated MPIOs. MagnReson Med 2011; 65: 1564-1569.
- Zhu XM, Wang YX, Leung KC, Lee SF, Zhao F, Wang DW, Lai JM, Wan C, Cheng CH, Ahuja AT et al. Enhanced cellular uptake of aminosilane-coated superparamagnetic iron oxide nanoparticles in mammalian cell lines. Int J Nanomedicine 2012; 7: 953-964.
- Al Faraj A. Preferential magnetic nanoparticle uptake by bone marrow derived macrophages sub-populations: effect of surface coating on polarization, toxicity, and in vivo MRI detection. Journal of Nanoparticle Research 2013; 15:1-13.
- Chen Z, Xu R, Zhang Y, Gu N. Effects of proteins from culture medium on surface property of silanes- functionalized magnetic nanoparticles. Nanoscale Res Lett 2008; 4: 204-209.
- Orts-Gil G, Natte K, Thiermann R, Girod M, Rades S, et al. On the role of surface composition and curvature on biointerface formation and colloidal stability of nanoparticles in a protein-rich model system. Colloids Surf B Biointerfaces 2013; 108: 110-119.
- Messaoudii C, Boudier T, Sanchez Sorzano CO, Marco S. TomoJ: tomography software for three-dimensional reconstruction in transmission electron microscopy. BMC Bioinformatics 2007; 8:288-294.
- Wolfgang Baumeister, Rudo Grimm, JochenWalz. Electron tomography of molecules and cells. Trends in Cell Biology 1999; 9: 81-85.
- Sorzano CO, Messaoudi C, Eibauer M, Bilbao-Castro JR, Hegerl R, Nickell S, Marco S, Carazo JM. Marker-free image registration of electron tomography tilt-series.? BMC Bioinformatics 2009; 10:124-131.